Jan Korbel
Head of Data Science
jan.korbel [at] embl.de
ORCID: 0000-0002-2798-3794
EditFrom genomic variation to molecular mechanism

Head of Data Science
jan.korbel [at] embl.de
ORCID: 0000-0002-2798-3794
EditGenetic variation studies have uncovered that genomic structural variants (SVs) such as deletions, insertions, and inversions account for most varying bases in human genomes. Recent studies indicate that somatic SVs occur post-zygotically throughout our lifespan, and show association with ageing and human diseases – calling into question the long-held belief that the genome is largely static within an individual, and preserved across all cells therein.
We employ a diversity of omics and imaging approaches – from single-cell multi-omics to spatial and bulk-cell omics as well as state-of-the-art microscopy – to investigate molecular mechanisms behind complex human phenotypes associated with genetic variants.
In addition to experimental methodologies applied to tissues and organoids, our laboratory is devising data science techniques including state-of-the-art machine learning methods for processing high-dimensional single-cell data sets, and for coupling genetic variation discovery with molecular and clinical phenotype data.
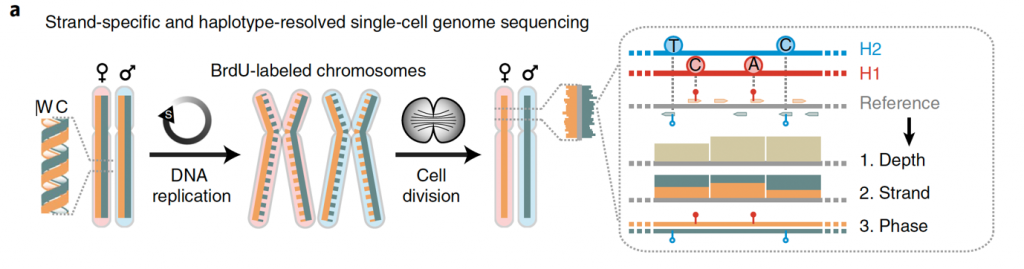
Of particular interest is understanding patterns of genetic mosaicism at cellular resolution. Our scTRIP method (Sanders et al., Nat Biotechnol 2020, Fig. 1) enables the direct detection of SV mutational processes in single cells, and as such can be used to obtain insights into pathomechanisms acting in human tissues.
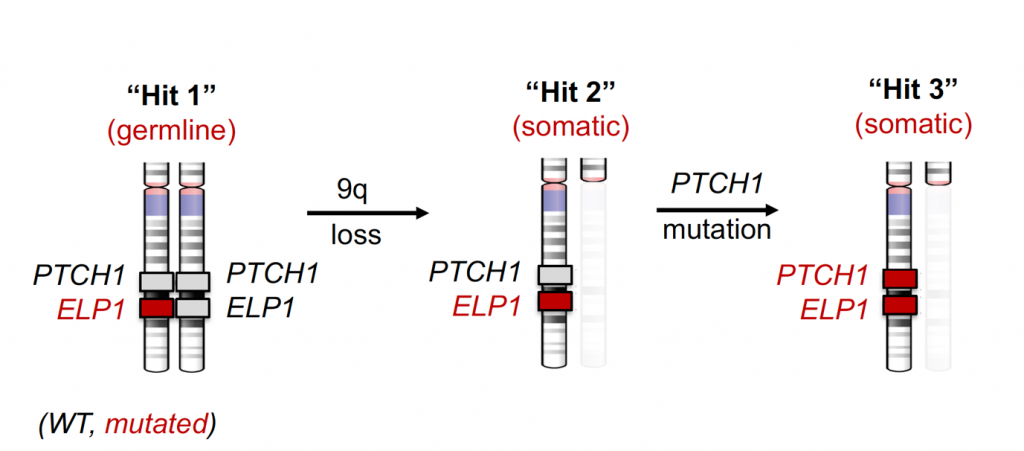
Another interest centres around uncovering commonalities and differences between molecular disease mechanisms in disparate cancer entities. In a rare-variant association study in medulloblastoma (MB) genomes/exomes, we recently described rare germline loss-of-function variants in the Elongator complex protein 1 (ELP1) gene in 15% of childhood MB genomes driven by Sonic hedgehog signalling (Waszak et al., Nature 2020). ELP1-associated MBs exhibit somatic loss of the wild-type ELP1 allele mediated by somatic large deletions that concomitantly cause loss of the PTCH1 gene residing adjacent to ELP1 on chromosome 9, involving an intriguing ‘three-hit’ molecular process (Fig. 2).
With respect to data science, our group has pioneered the utilisation of cloud computing to enable the global sharing and processing of large-scale biological data. We also contribute to develope haplotype-resolved human genomes to promote genotype-phenotype (such as eQTL) mapping and to generate highly resolved maps of genetic variation (Ebert et al. Science 2021) to support precision medicine approaches. Our group is also actively involved in building the German Human Genome-Phenome Archive (GHGA), a national research data infrastructure for disseminating and federating human genomics data from German studies nationally and internationally.