Best short talk winners at New Approaches and Concepts in Microbiology
The popular symposium “New Approaches and Concepts in Microbiology” took place virtually this year. 598 people from across the globe joined from their own time zone. Two presenters impressed the crowd with their short talks, even though the local time for one of them was 4.50 am (that doesn’t count as morning yet, does it?).
Jordi van Gestel and Nitzan Tal were the well-deserved winners. Read about their research below.
Short-range quorum sensing controls horizontal gene transfer at micron scale in bacterial communities
Presenter: Jordi van Gestel, University of California, San Francisco (UCSF), USA
Introduction: I am a Postdoc in the laboratory of Carol Gross at UCSF. Being trained as an evolutionary biologist, I was introduced to the fascinating world of microbiology during my PhD and have been working at the interface of both fields ever since. My research focuses on the organisation and evolution of bacterial cell collectives.
Abstract
Inside bacterial communities, cells often communicate through the release and detection of small diffusible molecules, a process termed quorum-sensing.
In general, signal molecules are thought to broadly diffuse in space; yet, paradoxically, cells often employ quorum-sensing to regulate traits that strictly depend on the local community composition, such as conjugative transfer. This raises the question if and how nearby cells in the community can be detected.
Here, we employ a microfluidic platform to determine how diverse quorum-sensing systems, differing in their regulatory design, impact the range of communication. While some systems indeed support long-range communication, we show that other systems support a novel form of highly localized communication.
In these systems, signal molecules propagate no more than a few microns away from signalling cells, due to the irreversible uptake of these signal molecules from the environment. This enables cells to accurately detect micron scale changes in the community composition and engage in local cell-to-cell communication.
Intriguingly, several mobile genetic elements, including conjugative elements and phages, employ short-range communication to specifically assess the fraction of susceptible host cells in their vicinity and adaptively trigger horizontal gene transfer in response. Our results underscore the complex spatial biology of bacteria, where cells both communicate and interact at widely different spatial scales.
Jordi is describing their @NatureComms paper published recently 👇 a microfluidic setup + amazing time lapse movies with measuring signal propagation of ComX, PapR, and PhrP – absorbing vs non-absorbing systemshttps://t.co/hQ6CfGVput#EESMicrobiology pic.twitter.com/RUtPEe0ZxY
— Akos T. Kovacs (@EvolvedBiofilm) July 8, 2021
Antiviral defense via nucleotide depletion in bacteria

Presenter: Nitzan Tal, Department of Molecular Genetics, Weizmann Institute of Science, Israel
Introduction: I am a PhD student in the lab of Professor Rotem Sorek at the Weizmann Institute of Science. For the past few years I’ve been studying the interactions between bacteria and their viruses (bacteriophages), and how both adapt to ever changing conditions in order to survive. My research focuses on identifying novel anti-viral defense systems and on understanding the extremely diverse arsenal of microbial immunity.
Abstract
DNA viruses and retroviruses need to consume large quantities of deoxynucleotides (dNTPs) when replicating within infected cells. The human antiviral factor SAMHD1 takes advantage of this vulnerability in the viral life cycle, and inhibits viral replication by degrading dNTPs into their constituent deoxynucleosides and inorganic phosphate.
In this study, we report that bacteria employ a similar strategy to defend against phage infection. We found a family of defensive dCTP deaminase proteins that, in response to phage infection, convert dCTP into deoxy-uracil nucleotides. A second family of phage resistance genes encode dGTPase enzymes, which degrade dGTP into phosphate-free deoxy-guanosine (dG) and are distant homologs of the human SAMHD1.
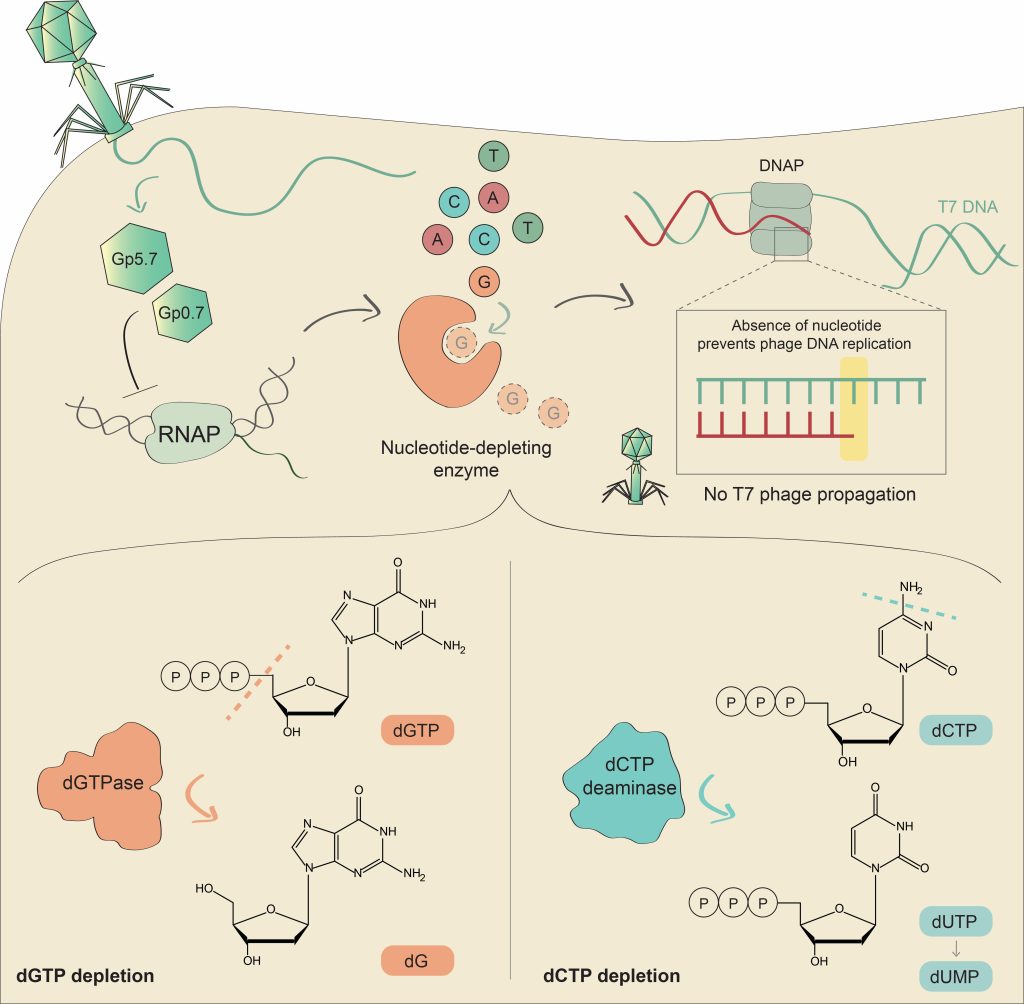
Our results show that the defensive proteins completely eliminate the specific deoxynucleotide (either dCTP or dGTP) from the nucleotide pool during phage infection, thus starving the phage of an essential DNA building block and halting its replication. Our study demonstrates that manipulation of the deoxynucleotide pool is a potent antiviral strategy shared by both prokaryotes and eukaryotes.
For tips and tricks on how to give a good scientific talk, watch this video.
