The rise of GPU computing in science
Discover how EMBL scientists are using GPU computing to push biology forward
By Berta Carreño and Laura Howes
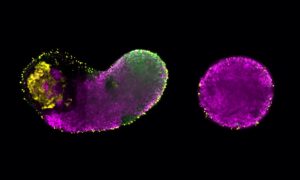
A couple of years ago, researchers at EMBL Barcelona did something quite radical. They threw away their carefully crafted software and started again from scratch. The reason, indirectly, was computer gaming.
“I had discussed it with my team on and off over the past five years,” says James Sharpe, head of EMBL Barcelona. “But there is a lot of effort involved in rewriting or writing a new simulation from scratch.”

It took the right person to come along before the Sharpe group could take the plunge. When Philipp Germann joined the group as a postdoc, he played with the existing software for a couple of months before deciding to completely rewrite a multicellular dynamics simulation, this time designed to run on graphics processing units (GPUs). The resulting new software can run simulations with hundreds of thousands of cells in a matter of seconds on a single graphics card. The previous software would take minutes, hours or even days to run. In the words of Sharpe, “It ended up fantastically well.”
Graphical computation
Open up any personal computer and you will find it packed with chips and components. They include the central processing unit (CPU), which does a lot of the heavy lifting and complicated processing, and the GPU which quickly creates the images you see on your screen – such as this article you are reading, the ad in your Facebook feed or the computer game you play at the end of the day to relax. GPUs contain hundreds or thousands of very specialised processors called cores. These cores are small and simple, and although they’re not as flexible as CPU cores, they can do one particular thing very, very fast: work out what your screen should display.
In computer games, every little region of the screen is the result of an independent mathematical calculation that works out what that little bit of the screen should look like. You can do those calculations one by one, or you can divide the screen into lots of little bits, and do all the calculations in parallel. That’s exactly what the GPU is good at: doing lots and lots of identical calculations simultaneously, with each one independent of the others. As computer game graphics became more complex – from Spacewar to Call of Duty – so did GPUs. The problem of how to render the image from a computer game is split into hundreds of thousands of little parallel calculations all done in a fraction of a second.
What is GPU computing?
The central processing unit (CPU) is the ‘brain’ of a computer. Its function is to carry out calculations that enable the computer to run software. The CPU is split into processing units, called cores, that receive instructions, perform calculations, and take actions on these instructions. The key power of the CPU is its flexibility to multitask with many different types of jobs. It can perform tasks quickly but, like our conscious brain, can focus on only a few ‘threads’ at a time.
The graphics processing unit (GPU) is a specialised component that was designed to handle graphics. Whereas CPUs have from four to eight cores, GPUs consist of thousands of small cores that can handle many threads simultaneously.
GPU computing is the application of GPUs to accelerate the CPU’s computing by transferring compute-intensive portions of the code to the GPU, where many threads can be handled in parallel. For suitable tasks, this makes calculations much faster to perform and offers cost and power efficiencies.
GPU computing was initially developed for graphics-intensive computational problems such as 3D rendering and gaming, but is now being applied to a variety of domains including complex modelling, simulation and cutting-edge research – such as the Sharpe group’s computer simulations of mammalian limb development.
“For us,” explains Sharpe, “it turns out that the kind of calculation required for computer games is similar to the kind of calculation we want to do. We try to simulate tissues and organs, how they grow, how their development works. And similarly, if we have a tissue with a hundred thousand cells, we can divide that cell population into little groups of cells, and each GPU can do the calculation for that little group.” Just as the image on a screen can be divided up, so too can the model tissue. “Every cell has the same genome and every cell has to make the same calculations, that’s why it fits so well into GPUs.”
GPU software requires programming languages with extra features that deal with the parallelisation of the problem, such as CUDA, but “programming and writing simulation software is similarly complicated whether you are running it on CPUs or GPUs,” adds Sharpe. It can be challenging to find a programmer who can understand the biological questions and then write the code, he notes. Sharpe says that there’s one major benefit to this shift to GPU computing, though: cost. A full-size CPU cluster costs a lot of money and resources to run, he explains. “We have switched over to being able to run everything in our own lab on our own computers’ graphics cards. We will probably start using GPU clusters [racks of dedicated GPUs] in the future. But, still, it’s saving a huge amount of time and money in this work.”

Clusters of computation
In EMBL Heidelberg, the server room hums and flashes with computing power. Inside, the High Performance Computing (HPC) cluster serves scientists across EMBL’s sites and packages up their different problems to be solved by a mix of CPUs and GPUs.
“When I came here, there were no GPUs installed,” explains Jurij Pečar, the engineer who looks after the HPC cluster. “So I went around interviewing scientists to learn what they’d need. One of the main requests was that they had software coming up able to use GPUs, so we invested in our first machine. As people started using GPUs, they realised how it speeds up their work and then, of course, we had to buy more.”
That is hardly a surprise when you realise that the microscopy image analysis that used to take about 30 days to run on 250 CPUs now takes just 30 hours on a single GPU. And some of the largest users of the GPUs at EMBL Heidelberg are the microscopy facilities. “Only six years ago, we were still shooting film and we had to develop the film in the darkroom manually,” remembers Wim Hagen of the Electron Microscopy facility. “Develop, fix, wash, dry, then scan the negatives and hope you didn’t make a mistake. Good people could do three boxes a day. Nowadays, it’s fully automated. We get 2000 to 3000 images a day and that pushes things.”
Balancing the users
Just as microscopy technology has improved, so everything has scaled, requiring more and more computing power. But while it might seem intuitive that processing microscopy images, or modelling cells as if they were areas on a screen, could be suited to GPU computing, other teams have other uses for the HPC cluster – for example, using deep learning to process huge sets of data from cancer patients.
“Deep learning is a big buzzword and I’m also into it,” explains Esa Pitkänen, a postdoctoral fellow in the Korbel group at EMBL Heidelberg. “Graphic processing runs on linear algebra and while linear algebra is very straightforward mathematics, you need to do a lot of it. GPUs parallelise this computation on a massive scale, and that happens to be exactly the same mathematics that you use to train deep neural networks. It’s very simple: it’s a perfect fit.”
Animation shows how a deep-learning model learns to recognise sequence patterns that underlie cancer mutations. EMBL/Esa Pitkänen
Pitkänen’s data is based on several thousand tumour samples, and the data he has is multi-layered. “We have sequencing data, methylation data, transcriptomics data, and then auxiliary and clinical data to top it off.” Pitkänen is trying to get his program to recognise patterns by letting the software learn how to recreate the data. In a sense, being able to recreate the data from a simple code indicates that the code – a few numbers, for example – captures the essential patterns in the data. It could be considered a more refined version of the suggested purchases we all get when we go internet shopping: people who have this mutation here, and this attribute here, also share similar tumour characteristics. To do that on CPUs, says Pitkänen, would take tens to hundreds of times as long. Instead, he trained his model in two days. “The whole process of training the deep neural networks is inherently massively parallel. And the way GPUs work is really well suited to train those models,” Pitkänen explains.
Back at the HPC cluster, Pečar’s main challenge, he says, is working with the users so that the big jobs from the microscopes can run at the same time as other programmes like Pitkänen’s. According to Pečar, GPU users need to work out how to move data from the main memory to the GPU memory efficiently. This means that developing the algorithms for the GPU is getting more technical, which in turn is now driving innovation. “I think programming for games is more exciting in the short term,” he admits. “But I’d rather work on something that has at least the hope of making the world a better place some time in the future.”
Growing graphically
Although computer games first drove the development of GPUs, chip manufacturers are now optimising their GPUs for other applications, like Sharpe’s modelling or Pitkänen’s deep-learning applications. It seems GPU-based computing is growing in the life sciences, and in the EMBL IT department.
“There are parts of biology that are not so suited to GPU computing, like some tasks in sequence-based informatics where you’re first uploading a huge dataset, and then slowly chugging your way through it,” concludes Sharpe. “But I believe in a certain version of systems biology – the approach in which you build computer simulations of a biological process as a way of understanding the dynamical mechanisms of the system. Using computers in this way – simulating dynamics [rather than analysing data] – is still quite rare, but I expect and hope it will grow and become a part of all biological projects.”


