
Peer Bork
Interim Director General
EditUnderstanding life in its natural context
Searchable Planetary-scale mIcrobiome REsource.
SPIRE integrates metagenome-derived microbial data modalities across habitats, geography and phylogeny.
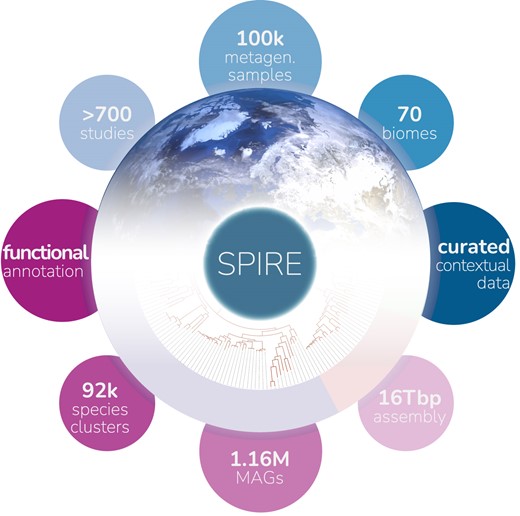
SPIRE is a recently published platform with the aim to provide an integrated view of microbial diversity at various taxonomic granularities, at genome and at gene level, across known microbial habitats. It is a “one-stop-shop” for microbial data, integrated and consistently processed across habitats and phylogeny, at global scales. SPIRE is derived from ~100k metagenomic samples in 739 studies encompassing ~500Tbp of publicly available raw metagenomes, and holds five main types of data:
Publication
Nucleic Acids Research, Volume 52, Issue D1, 5 January 2024, Pages D777-D783 https://doi.org/10.1093/nar/gkad943
Coelho, Queensland University of Technology
Schmidt, University College Cork