
Gautam Dey
Group Leader
Dey Group
EditEvolutionary cell biology of the nucleus

Group Leader
Dey Group
EditThe nucleus is an immensely complex structure at the heart of every eukaryotic cell: gated by nuclear pores, scaffolded by a laminar mesh, and tethered to the genome. It is also highly dynamic – each cell cycle, the entire nucleus is rapidly remodelled in perfect lockstep with cell division. Defects in the nuclear compartment, or its ability to remodel, have dramatic consequences for genome integrity and cell survival. Yet different species have evolved vastly divergent ways to organise their nuclei – for example, some completely dismantle their nuclear envelope during mitosis, while others keep it intact. How and why did these different strategies emerge, and do the nuclei of extant eukaryotes retain any shared, fundamental organisational principles? Could understanding these core mechanisms help us understand nuclear (dys)function better?
These questions motivate our research. We recently uncovered the mechanism by which fission yeast nuclei divide (Fig. 1) – a process that exhibits unexpected parallels to nuclear remodelling in animal cells (Dey et al., Nature 2020). We have also investigated the archaeal origins of the nucleus, using experimental models (Tarrason Risa et al., Science 2020; Pulschen et al., Current Biology 2020) and the proteomes of uncultured archaea (Dey et al., Trends Cell Biol 2016). Our work suggests for the first time that deeply conserved mechanisms might unite nuclear remodelling strategies once thought to be distinct.
In earlier work, we developed a novel phylogenetic method to detect protein co-evolution. This work revealed ancient, co-evolving modules in the human genome that we could study experimentally (Dey and Meyer, Cell Syst 2015; Dey et al., Cell Rep 2015).
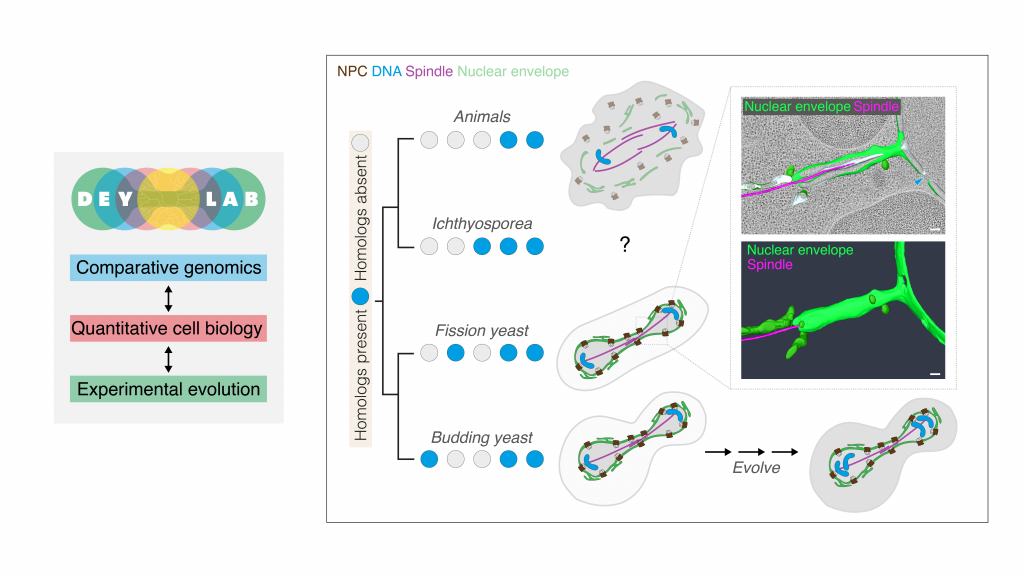
Our group at EMBL will employ an interdisciplinary toolkit (microscopy, genomics, gene editing) in a range of model systems (the fission yeast S. pombe, the slime mould Dictyostelium, and the apicomplexan parasites Plasmodium and Toxoplasma), to investigate the core mechanistic principles of nuclear organisation and dynamics (Fig. 2). Our comparative approach will help us better understand nuclear (dys)function while helping improve models of eukaryotic evolution. To highlight a couple of projects:
We have shown that S. pombe is a powerful, general purpose model for studying nuclear dynamics (Dey et al., Nature 2020). We will use it now to investigate how the nucleus and cell cycle control machinery communicate to ensure that copies of the genome are safely encapsulated before the cell divides. We will further study the dynamics of nuclear pore complexes throughout the fission yeast cell cycle, both their intensive remodelling during mitosis but also their homeostasis throughout interphase.
In all eukaryote, proper partitioning and genomic reorganisation is essential for successful division. However, the organisation of genomes – e.g. size and number of chromosomes – can vary immensely between organisms. We will investigate how cells are coping and adapting to having extra genomic material by studying changes in cellular dynamics and using experimental evolution (led by the Sherlock Lab).
Many protists, including a set of key human parasites and Ichthyosporea, have unusual and understudied nuclear envelopes, pores and cell cycles. We will extend our investigation to new experimental models – merging our findings into a phylogenetic map of molecular signatures for modes of nuclear organisation. By studying mitosis in the multinuclear state of Ichthyosporea in close collaboration with Jannik Schwab and Omaya Dudin, we aim to gain a deeper understanding on how strategies of nuclear organisation evolved.