Justin Michael Crocker
Group Leader
ORCID: 0000-0002-5113-0476
EditGene regulation during evolution and development

Group Leader
ORCID: 0000-0002-5113-0476
EditThe goal of my research group is to reach a level of understanding that will allow the programming and control of developmental fates, extending existing single-cell synthetic biology techniques to whole-organism developmental systems. To a large degree, development is controlled by regions of genomic DNA called enhancers that encode binding sites for transcription factor proteins. The binding of activators and repressors increases and reduces gene expression levels, respectively. Still, how combinations of activators and repressors generate precise transcription patterns during development is not understood. Towards this aim, we have developed high-throughput methods to dissect transcriptional enhancers to understand their role in determining gene expression patterns in the fruit fly, a classic model system. We explore the role of enhancer architecture on gene expression by creating synthetic networks for pattern formation. Our work uses new genome engineering technologies (stay tuned), synthetic biology (Galupa et al., 2023), high-throughput robotics (Fuqua et al., 2020), and experimental evolution (Li et al., 2024), creating a cross-cutting approach to developmental systems biology.
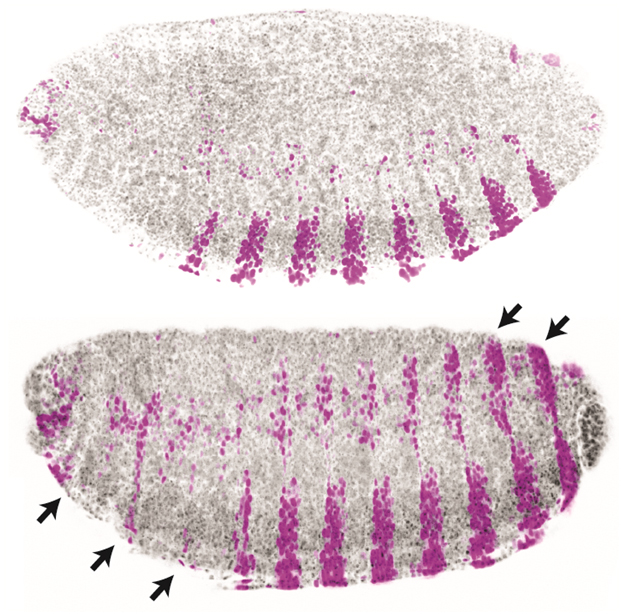
Enhancers are fundamental to development and evolution (Fig. 1), yet we lack a detailed understanding of their necessary components and how these components function collectively. We aim to determine the rules for designing and controlling synthetic enhancers—including the number, affinity, and positioning of binding sites for transcription factors. We will explore this problem using a fully synthetic transcriptional platform in fruit flies consisting of engineered transcription factor gradients and artificial enhancers.
We aim to combine quantitative models of enhancer function with manipulations using engineered transcription factors to examine how much enhancer function can be controlled predictably. We aim to develop models that allow the predictable control of enhancers, providing a framework for quantitatively controlling enhancers with engineered transcription factors.
Transcriptional networks contain the information required to confer robust positional information within a developing embryo. However, these networks have remained recalcitrant to modeling, as every parameter, such as diffusion and binding coefficients, are usually fitted values and not derived from in vivo data. Building fully synthetic networks allows for exploring network architecture unobscured by unknown elements. We aim to explore the role of network architecture on gene expression by creating synthetic networks for pattern formation.
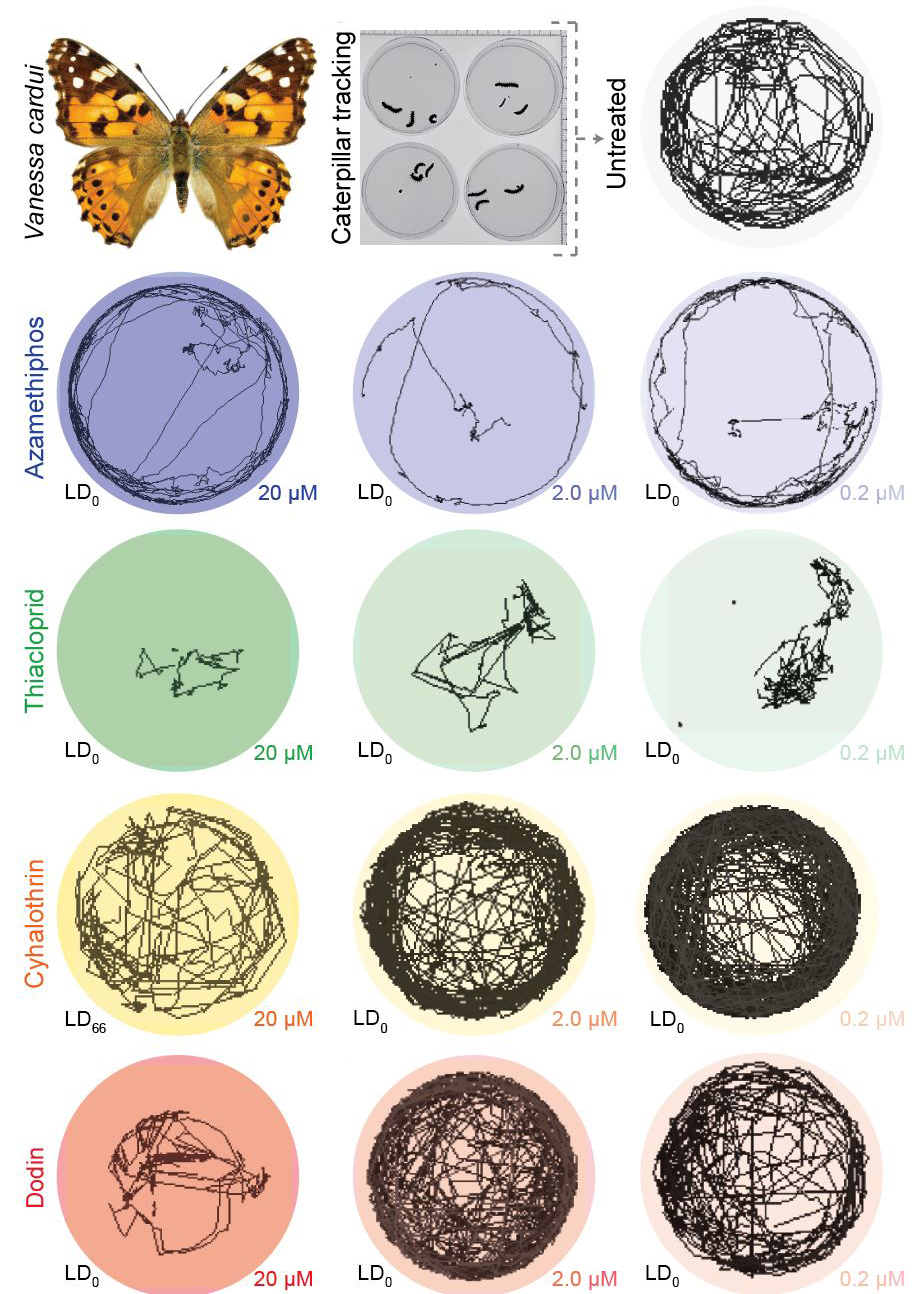
Insect biomass is declining across the globe at an alarming rate. Climate change and the widespread use of pesticides have been hypothesized as two underlying drivers. However, the lack of systematic experimental studies across chemicals and species limits our causal understanding of this problem. Using a controlled laboratory pipeline for Drosophila melanogaster, we recently found that many agricultural chemicals affect behavior at sublethal concentrations, and an even higher proportion compromises long-term survivability after acute exposure (Gandara et al., 2024). Finally, we expanded our investigation to additional fly species, mosquitos, and butterflies and detected similar behavioral alterations triggered by pesticides at sublethal concentrations (Fig. 2). Our results provide experimental evidence that suggests sublethal doses of agrochemicals coupled with changes in environmental temperatures significantly contribute to the global decline in insect populations. We anticipate that our assays can improve chemical safety assessment, better protect the environment, secure food supplies, safeguard animal and human health, and understand our rapidly changing world.
There are several major limitations of using fruit flies for experimental evolution: 1) there is a lack of sufficient genetic material to sample a broad range of genotypic space for pathways of interest; 2) the long generation times compared to microbial evolution experiments; and 3) while it is possible to generate diversity in the lab using EMS mutagenesis, a significant shortcoming of this approach is that it can lead to many background mutations (Li et al., 2024).
To facilitate large-scale experimental evolution, we have pioneered methods to increase targeted mutation rates in Drosophila, accelerating evolution and biasing it toward gene-regulatory networks of interest (Li et al., MBE 2024).
We are now combining high-throughput behavioral assays with directed evolution towards AI in the loop. Stay tuned…