Eileen Furlong
Head of Genome Biology Unit
ORCID: 0000-0002-9544-8339
EditGenome regulation and chromatin topology during embryonic development

Head of Genome Biology Unit
ORCID: 0000-0002-9544-8339
EditPrecise regulation of gene expression is essential for almost all biological processes, and a key driving force in development, evolution and disease. Expression states are initiated through diverse cues modulating transcription factor activity, which converge on cis regulatory elements called enhancers. Enhancers thereby act as integration platforms to control specific patterns of expression, telling genes when and where to be expressed. Given their central role, mutations in enhancers often lead to devastating developmental defects and human disease.
Our research focuses on dissecting general principles of genome regulation, including how the cis-regulatory genome is organised within the nucleus, and how chromatin state and transcription factors influence this process. Our work combines (single-cell) genomic, genetic, imaging and computational approaches to understand these processes, including the development of new genomic methods within the context of a multicellular embryo.
For enhancers to function, they must come in proximity to their target genes. We previously found that developmental enhancers are often in proximity to their promoters hours before gene expression (Ghavi-Helm, Nature, 2014). These preformed topologies are contained within chromatin domains called TADs. Using extensive rearrangements, we recently showed that TAD boundaries do not restrain enhancer function in the majority of cases, as generally thought. When a boundary is removed, most enhancers still regulate their appropriate target gene (Fig. 1; Ghavi-Helm, Nature Genetics, 2019). We are currently dissecting what role TADs play in gene expression using high-resolution imaging, genomics and genetics.
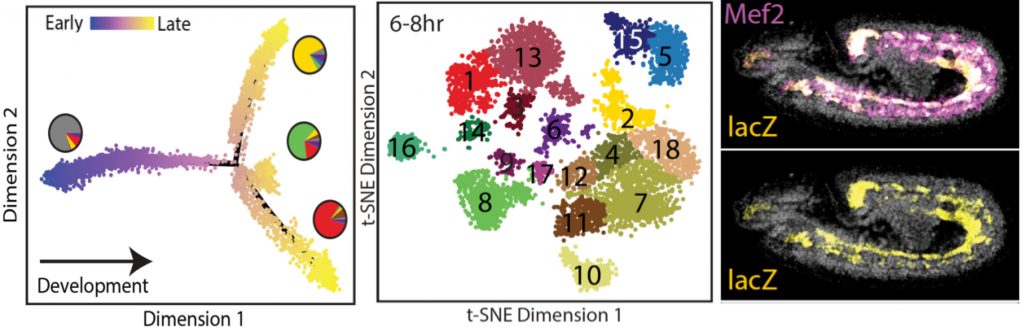
To uncover general properties of enhancer function we develop and optimise methods to investigate tissue specific (Bonn, Nature Genetics, 2012), and single-cell (Fig. 2; Cusanovich, Nature, 2018) changes in enhancer usage during embryonic development. Going forward, we will combine single cell methods with CRISPR/Cas9 and optogenetics to functionally dissect regulatory properties associated with developmental transitions.
Variation in the non-coding genome is widely associated with quantitative traits. Many disease-associated variants are in cis-regulatory elements, yet it is very difficult to pinpoint the causal SNP within human genomes and dissect their underlying mechanism. We recently demonstrated that we can bridge this gap using drosophilids, thanks to their small blocks of LD (Cannavò, Nature, 2016; Schor, Nature Genetics, 2017). We are using natural sequence variation as a perturbation tool to developmental programming to uncover inherent mechanisms that impart robustness, or canalisation, within developmental networks.