Instrumentation
Available systems at GenTechDev
Custom multi-omics tech development
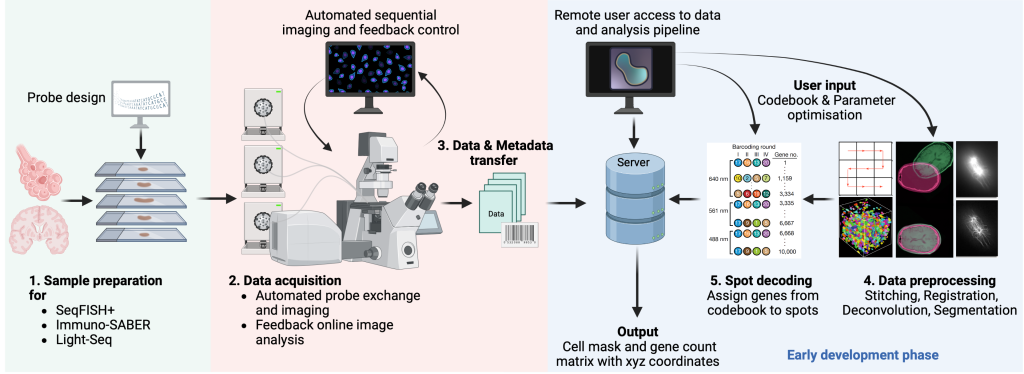
SeqFISH+ is a single molecule FISH approach to visualize RNA molecules in tissues. Using sequential rounds of imaging, gene specific barcodes can be demultiplexed by hybridizing short fluorescent imager oligos. Increasing the imaging rounds allows to increase the number of barcodes so that seqFISH+ can be scaled up to the whole transcriptome. In addition, seqFISH+ can be combined with sequential immunofluorescence to integrate protein readouts.
Eng, CH.L., Lawson, M., Zhu, Q. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019). https://doi.org/10.1038/s41586-019-1049-y
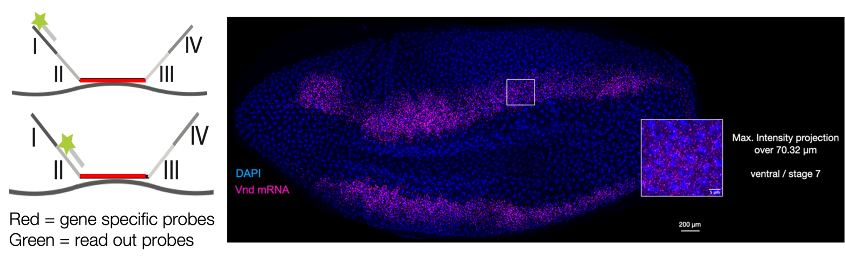
Immuno-SABER is a technique for multiplexed and sensitive in situ protein detection with individually programmable signal amplification. Tissue samples are labelled with antibodies conjugated to DNA-barcodes, which are hybridized to long single-stranded concatemeric sequences. These sequences act as scaffolds for >100-fold signal amplification of the antibody signal, when bound to fluorescent imager probes.
In the process, multiplexed antibody panels are first applied to the sample, followed by one-step concatamer hybridisation, and rapid sequential hybridisation of fluorescent imaging oligos.
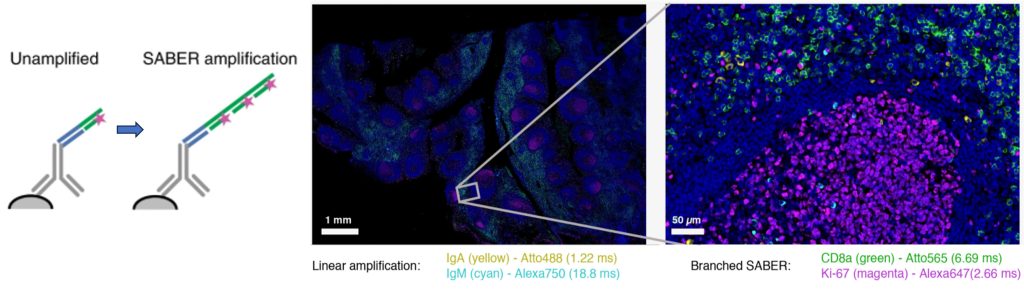
Figure 2. Left, schematic of SABER signal amplification. Right, Immuno-SABER example on a human tonsil FFPE tissue section
Saka, S.K., Wang, Y., Kishi, J.Y. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat Biotechnol 37, 1080–1090 (2019). https://doi.org/10.1038/s41587-019-0207-y
In-situ sequencing (ISS) is a powerful method of direct mRNA sequencing in large, fixed tissue sections at high resolution. Based on the rolling circle amplification (RCA) of padlock probes, it produces enough signal to be detected with a standard epifluorescence microscope and a 20X objective. Using a color-coding scheme and a few rounds of hybridization cycles, ISS can identify dozens to hundreds of target genes (~300) at single cell resolution. We are applying ISS to human and mouse tissue but the technique is applicable to any species with available transcriptome data.
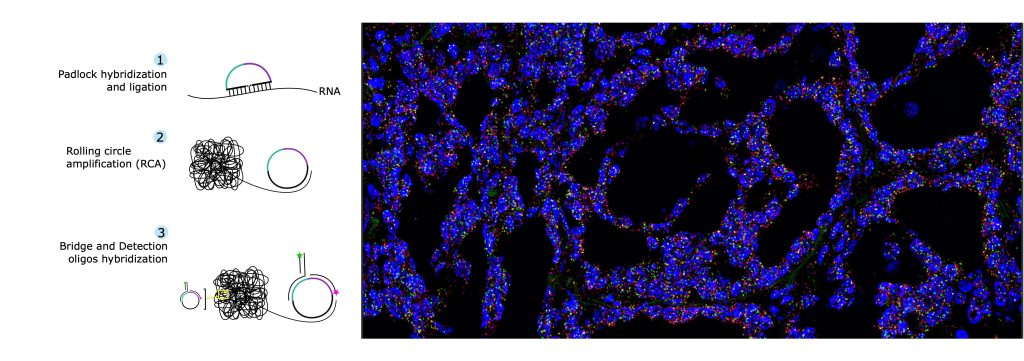
Lee, H., Marco Salas, S., Gyllborg, D. et al. Direct RNA targeted in situ sequencing for transcriptomic profiling in tissue. Sci Rep 12, 7976 (2022). https://doi.org/10.1038/s41598-022-11534-9
Available systems at GenTechDev