Marco Marcia
Visiting Group Leader (Outgoing)
ORCID: 0000-0003-2430-0713
EditStructure and function of lncRNA-protein complexes regulating development and stress responses

Visiting Group Leader (Outgoing)
ORCID: 0000-0003-2430-0713
EditMy group studies the molecular mechanism of mammalian lncRNAs involved in gene expression regulation during development and in response to environmental stress. We are particularly interested in how lncRNAs fold into discrete secondary and tertiary structures and how these structures determine cellular functions in association with regulatory proteins, such as chromatin remodeling enzymes and transcription factors.
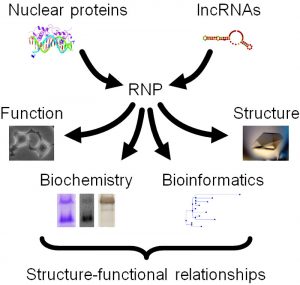
While we primarily use biochemical approaches, such as SHAPE and HRF probing, in combination with state-of-the-art biophysical methods, such as X-ray crystallography, cryo-electron microscopy, small angle scattering, atomic force microscopy, and NMR, we also crucially integrate these in vitro studies with in vivo functional assays, such as gene expression and cell cycle analysis (Figure 1).
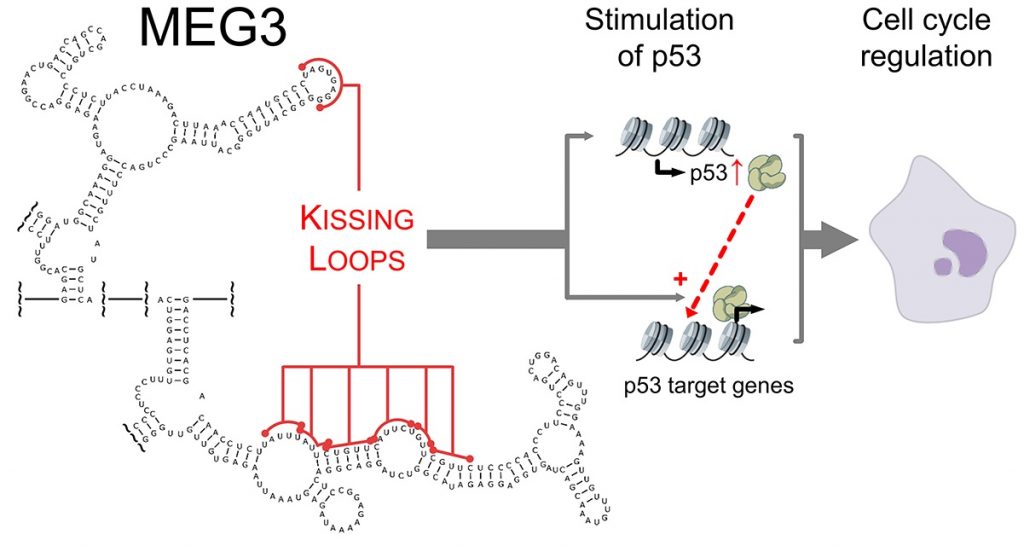
In a recent work, we have determined novel and unexpected secondary and tertiary structure properties of an imprinted alternatively-spliced human lncRNA named MEG3 (maternally expressed gene 3). Thanks to a systematic mutagenesis approach coupled to high-throughput in vivo functional assays and evolutionary alignments, we have surprisingly discovered that MEG3 needs to adopt a discrete tertiary structure to be able to stimulate protein p53, thus acting as a tumor suppressor in adult cells (Figure 2). In parallel, we are producing known interactors of MEG3, such as Polycomb group proteins and p53 itself, to characterize these proteins and their interactions with MEG3 from a biochemical and biophysical stem point.
Our current achievement now open the way for characterizing the structure and function of MEG3 and of many other lncRNAs at greater level of precision, by high resolution biophysical techniques and proteomics, transcriptomics, and genomics approaches. Our goal is to decipher at least some of the puzzles and mysteries surrounding the biology of lncRNAs. What are the molecular determinants for the apparently promiscuous, but tight and selective interaction of lncRNAs with their protein cofactors? What controls lncRNA folding in the crowded nuclear environment? What role does the complex structure of lncRNAs play in orchestrating the interplay of all epigenetic regulators and the structure of chromatin? Ultimately, obtaining high resolution insights into lncRNAs and their ribonucleoprotein complexes will allow to measure epigenetic processes quantitatively and potentially lead to the development of new therapeutic approaches to cure invasive human diseases.