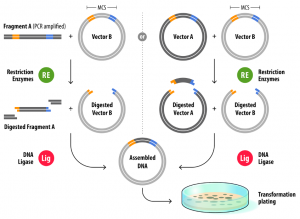
For classical restriction/ligation cloning, 2 types of enzymes (restriction enzymes and DNA ligase enzymes) are commonly used. Restriction endonucleases are enzymes that perform a sequence-specific cleavage of a DNA phosphodiester bond, while DNA ligases join DNA fragments together via the formation of a phosphodiester bond.
The start of restriction/ligation cloning is either a fragment amplified by PCR, in which your gene of interest is flanked by the chosen restriction sites, or a vector that already contains your gene of interest flanked by the chosen restriction sites. The vector you want to clone your gene of interest into should of course also possess the same restriction sites. You then digest both your PCR fragment (or vector containing your gene of interest) and vector with the same set of restriction sites. Several commercial providers offer “fast” versions of restriction enzymes, that can complete the reaction in 5-15 min in a universal buffer compatible with many different restriction enzymes, which significantly speeds up the cloning process. After the restriction enzyme reaction, you can clean up your digested PCR fragment and vector either by purification from an agarose gel after electrophoresis or directly using a commercial PCR purification kit. For the ligation reaction, you mix the digested vector and fragment in a ratio usually ranging from 1:2 to 1:4 (starting from approximately 50 – 100 ng of vector), after which you transform the ligation mixture into a competent E. coli cloning strain.
The advantage of restriction/ligation cloning is that there are more than 200 different restriction enzymes that are currently commercially available. Many expression vectors also have dedicated multiple cloning sites that can be used for restriction/ligation cloning. Unfortunately, not all vectors have compatible multiple cloning sites and therefore one cannot always simply “cut and paste” a fragment from one expression vector into another one. In contrast, the pETM vector series, which was developed at EMBL, is designed in such a way that it can be used to generate expression constructs of a gene of interest with a multitude of different tags in parallel starting from the same digested PCR fragment.
A disadvantage of restriction/ligation cloning is that the gene of interest might already possess several internal restriction sites, which excludes these for usage in classical restriction/ligation cloning. Finally, “seamless” or “scarless” cloning is also not always possible, unless one uses type II restriction enzymes, which cleave outside their recognition site. Golden gate cloning, for example, is based on the usage of type II restriction enzymes and does allow for seamless cloning. Vectors compatible with Golden gate cloning are nowadays also commercially available or via non-profit plasmid repositories such as Addgene.