Sinem K. Saka
Group Leader
sinem.saka [at] embl.de
ORCID: 0000-0002-5017-379X
EditSpatial biology from molecules to tissues: high-dimensional investigation of cellular organisation

Group Leader
sinem.saka [at] embl.de
ORCID: 0000-0002-5017-379X
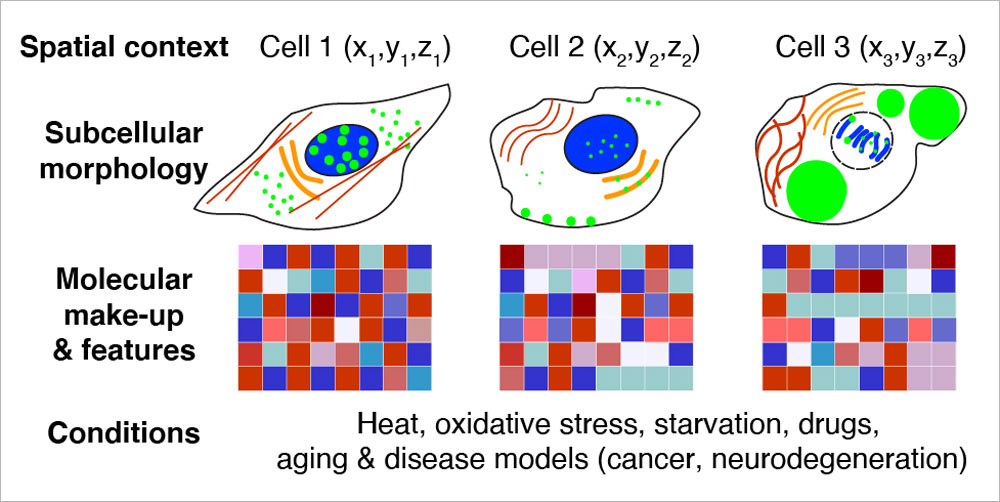
EditWe aim to understand the spatial features and organisational principles of biological structures ranging from nanoscale complexes to subcellular compartments, cells, and tissues. To enable comprehensive investigation, we develop molecular tools and methods for conventional and super-resolution imaging, and employ multimodal approaches.
Previously, we have combined metabolic labelling, bioorthogonal click chemistry, cell fusion assays, and super-resolution microscopy to understand the universal principles of protein organisation in cellular membranes. We have also developed a correlative optical and isotopic super-resolution imaging modality (COIN) that makes it possible to study the turnover of proteins in subcellular compartments of neurons.
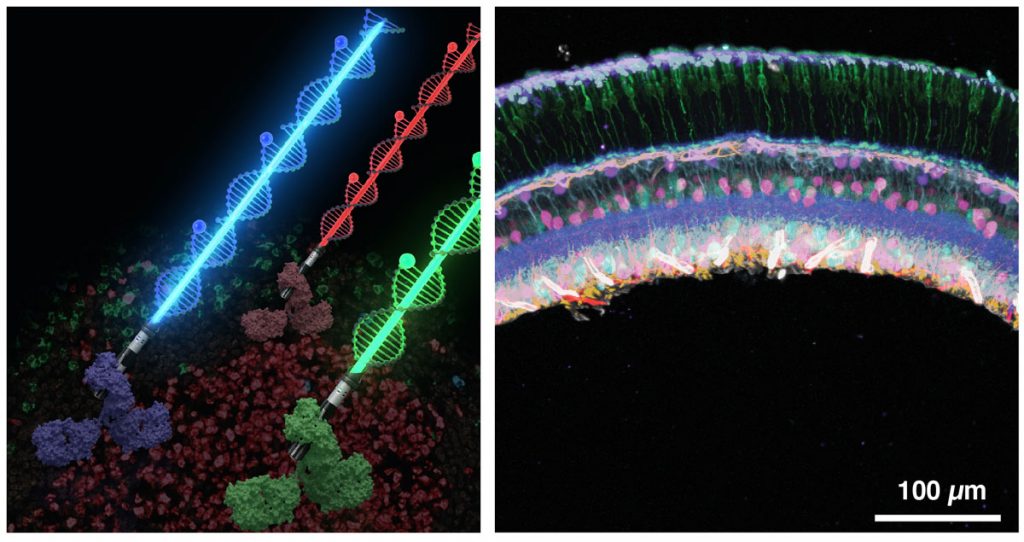
Recently, we have harnessed the barcoding capacity and programmability of DNA to generate powerful tools for single-molecule imaging and multiplexed detection of proteins and nucleic acids in cells and tissues. We are expanding and applying these tools to visualise the spatial organisation and heterogeneity of subcellular structures and organelles.
To broaden DNA-based detection further to the tissue scale, we have developed a new signal amplification method called signal amplification by exchange reaction (SABER) for immunofluorescence (Immuno-SABER) and fluorescence in situ hybridisation (SABER-FISH) to enable high-throughput multiplexed profiling of single cells in their native tissues. We are currently using SABER to reveal the spatial features of cellular functions in human tissues.