
Yannick Schwab
Team Leader and Head of Electron Microscopy Core Facility
ORCID: 0000-0001-8027-1836
EditVolume correlative light and electron microscopy

Team Leader and Head of Electron Microscopy Core Facility
ORCID: 0000-0001-8027-1836
EditOur team is developing methods that aim to describe the subcellular organisation of cells in their native context. Almost all biological systems bear a complex organisation that makes it challenging to target electron microscopy observations to single events or cell types. Multimodal correlative imaging appears now as a powerful solution to efficiently navigate the complexity of such specimens. Whilst focusing our efforts on the imaging techniques, we are developing strong collaborations with laboratories whose research in Biology is confronted with the challenge of capturing these rare events at high resolution. Together, we strive to find adapted solutions and then make them accessible to the community. As a consequence, our laboratory is involved in many fascinating fields of Life Sciences, including cancer research, host-pathogen interactions, neurosciences, evolution, cell and development biology.
Thanks to our tight links with the EMBL’s Electron Microscopy Core Facility, we are using a large portfolio of sample preparation methods, EM modalities such as TEM, TEM tomography, SEM and volume SEM (FIB-SEM and SBF-SEM) and image analysis tools. Our core activity is to develop methods in multimodal correlative imaging that cover Correlative Light and Electron Microscopy (CLEM), in 2D for Live-cell CLEM but also in 3D with Volume CLEM; Correlative Light, X-ray and EM (CLXEM) and Correlative X-ray and EM (CXEM) which are both best adapted to multicellular, rather bulky specimens.
One of the methodology approaches in the lab is the use of targeted light and electron microscopy to study methods for applications in cell biology processes. Here we work with various cell lines, focusing on developing strategies to image cellular processes in live and chemically fixed cells.
Our ultimate goal is to link the dynamic behaviour of a particular cellular event, and the molecular information, obtained by means of light microscopy with the ultrastructural details provided by EM. To achieve this goal, we apply volume EM methods, including FIB-SEM and SBF-SEM, enabling us to get further insights into the complex 3D organisation of the subcellular compartments. Furthermore, in order to get meaningful results, we aim at increasing the throughput at all stages of the workflow and, thus, at developing quantitative tools.
Previous work shows how Live-cell CLEM was used to capture cells at metaphase (Cosenza et al 2017), as well as our attempt to automate the acquisition of multiple target cells with a FIB-SEM (Lleti, Steyer, Schieber et al 2022).
Currently we have two main projects within this topic: one centered around immune cell migration, with the group of Alba Diz-Muñoz, and the other around regulation of cell division with the group of Anne Lore Schlaitz (Heidelberg University). These biological processes are both highly dynamic, Furthermore, the study of these processes implies that rare events need to be precisely located and targeted within a large population of cells. For these reasons and to fully understand these events, it is essential to correlate the information coming from both imaging modalities within the exact same cells: the link between the cellular history generated via live cell imaging with the ultrastructural information originated with EM imaging.
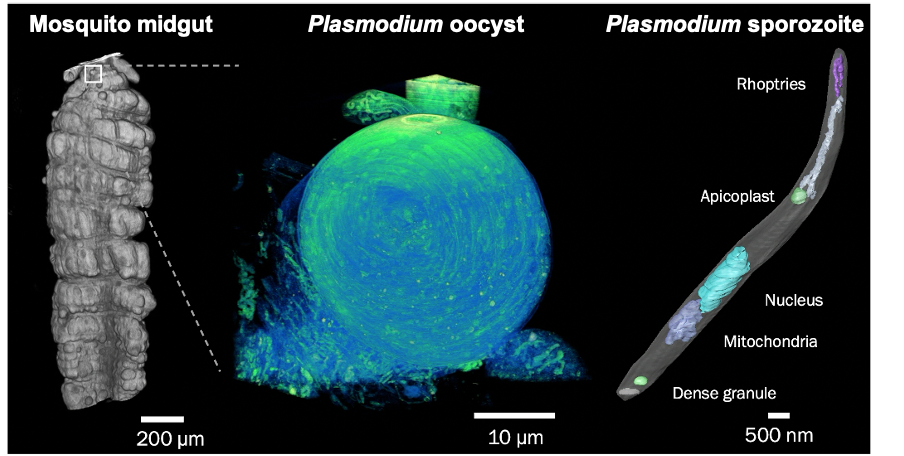
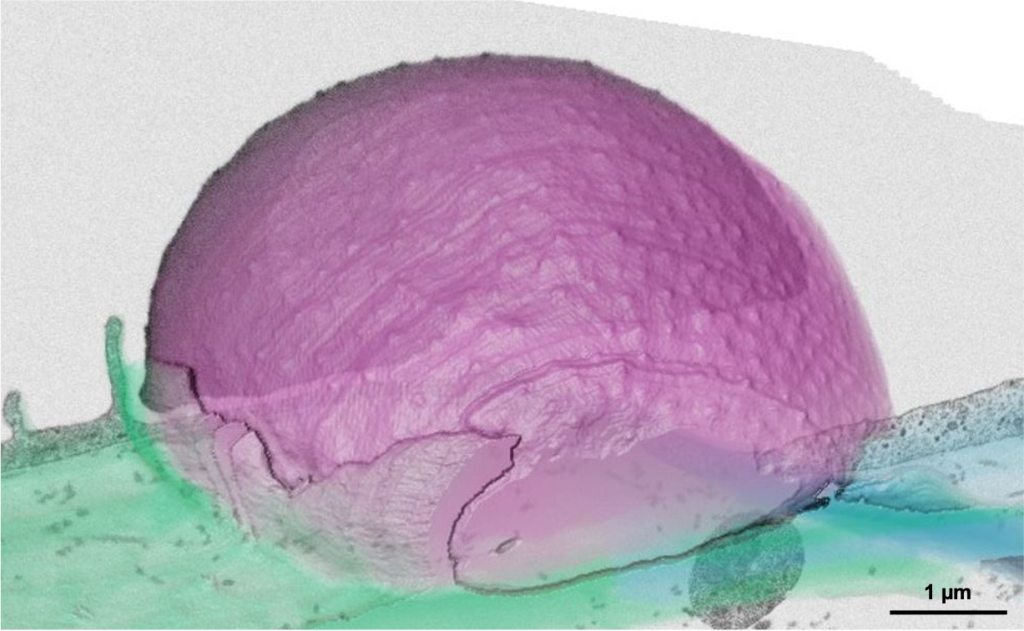
Malaria still poses a major threat to human health. The life cycle of the malaria parasite Plasmodium spp. is very complex and involves a host (humans and other vertebrates) and a vector (mosquito). Besides its medical importance, Plasmodium has a fascinating biology, undergoing enormous cell biological and physiological changes in the different life cycle stages. Despite the greatest efforts, scientists have not succeeded yet to unlock all of Plasmodium‘s cell biological secrets. This includes its development within the mosquito vector, as well as the pathological aspects of the infection. In order to further decipher the development of the malaria parasite, we have established a fruitful collaboration are collaborating with the group of Freddy Frischknecht at Heidelberg University at the Center for Integrative Infectious Diseases.
Furthermore, in tight collaboration with Maria Bernabeu at EMBL Barcelona we are also studying the pathogenesis of cerebral malaria, a severe and often lethal complication characterised by deep coma. Taking advantage of a bioengineered 3D brain microvasculature model, that mimics the blood-brain barrier (BBB), we aim at revealing detailed insights into the unknown mechanisms of how infected red blood cells (iRBCs) interact with the BBB, resulting in cerebral malaria.
For both malaria projects, we are developing correlative multimodal 3D imaging approaches using confocal microscopy, microCT (computed tomography), synchrotron-based X-Ray imaging (in collaboration with Liz Duke and Thomas Schneider at EMBL Hamburg), guided ultramicrotomy and volume SEM to obtain 3D ultrastructural details of target regions of interest.
We are particularly interested in applying advanced imaging techniques to study the fine ultrastructure of marine microorganisms. Ongoing collaboration with the group of Johan Decelle (University of Grenoble-Alpes, France) has shown the power of volume EM to unravel the ultrastructure of non-model, non cultivable protist symbioses (e.g. Uwize et al 2021). The high heterogeneity of microplankton in environmental samples makes it challenging to focus on selected species. Yet, increasing our knowledge about the morphology of these crucial components of the marine ecosystems is key to better understanding better their response to changing environments or environmental conditions. We are thus working on developing and/or applying multimodal correlative imaging techniques to analyse targeted taxa from mixed populations collected in the field. After 4 pilot expeditions conducted in 2019, 2020, 2021 and 2022 in Italy, France and Iceland, the team is now contributing to the – TREC (Traversing European Coastlines) expedition. By systematically sampling microplankton at several European marine stations, we plan to build an ultrastructural atlas of dinoflagellate biodiversity, linking morphological variations with ecological gradients.
Along these lines, we have recently developed a correlative approach (jointly with the EMCF and the ALMF) that allows the study of plankton biodiversity at a sub-cellular level (https://journals.biologists.com/jcs/article/136/15/jcs261355/325830/Targeted-volume-correlative-light-and-electron). This paper has attracted a lot of attention in the scientific community. Thus, it has been highlighted by the Company of Biologists (https://journals.biologists.com/jcs/article/136/15/jcs261499/325833/First-person-Karel-Mocaer) (https://journals.biologists.com/jcs/article/136/15/e136_e1502/325835/vCLEM-reveals-the-ultrastructure-of-phytoplankton) and made it to the cover of Journal of Cell Science. Leica Microsystems has also published an application note about this methodology (https://www.leica-microsystems.com/science-lab/authors/detail/karel-mocaer/).
Furthermore, this methodology is going to be covered in April 2024 during an EMBO lecture course (Imaging Marine Organisms Across Scales) at the Stazione Zoologica in Napoli (https://coming-soon.embo.org/lc24-01). This course is organised alongside a TREC stop at the Stazione Zoologica.
