Vikas Trivedi
Group Leader and Co-chair of Theory at EMBL Transversal Theme
ORCID: 0000-0003-0953-0553
EditSelf-organisation in multicellular systems

Group Leader and Co-chair of Theory at EMBL Transversal Theme
ORCID: 0000-0003-0953-0553
EditControl mechanisms and robustness of multicellular self-organisation
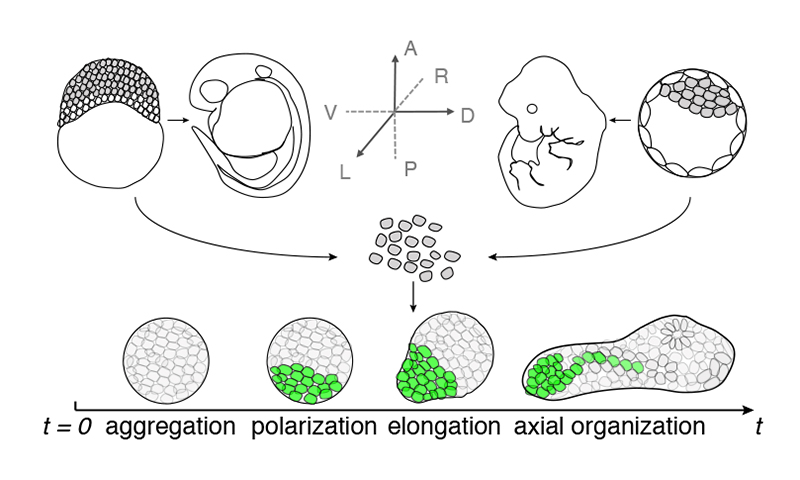
How do tissues reliably form complex shapes and patterns in multicellular animals? Despite a century of embryology, the integration of genetic networks, mechanical forces, and cellular metabolism in self-organisation remains a mystery, partly because it’s hard to untangle their combined effects. Using aggregates of embryonic stem cells from mouse, human, and other models, we recreate early developmental events in the lab. By probing how mechanochemical coupling between cells interacts with metabolic constraints, we uncover fundamental rules guiding robust symmetry breaking and tissue patterning during the earliest stages of development.
Evolutionary perspective on emergence of multi-cellular structures
Using pluripotent cells from various vertebrates, we found that removing species-specific geometric limits reveals a conserved morphogenetic program. By engineering glucose availability in mouse and human stem cells, we can control patterning through metabolism, influencing cell fate. Inspired by the role of metabolism in mammalian stem cell aggregates, we are looking into nutrient regulation of the cell cycle and explore how it could have contributed to the emergence of multicellularity in species such as Capsapora owczarzaki.
Modelling Immune Tissue Development
Building on our studies of early developmental self-organisation, we develop human iPSC-based 3D models to explore the generation of the lymphatic system in vitro. This bottom-up approach combines directed differentiation along lymphatic lineages in combination with endothelial, and mesenchymal cells to reveal the minimal requirements for multilineage tissue self-assembly, offering new insights into immune organ development and multicellular coordination.
Universal mechanisms of temperature resilience in embryos
Building on our studies of multicellular self-organisation, we explore how environmental challenges such as temperature extremes impact robust tissue development in ectothermic embryos like zebrafish. While thermal sensitivity of cellular chemical reactions can be the (trite) reason, it’s not intuitive how mechanics (tissue rheology, movement) and chemistry (morphogens) coordinate when temperature changes. Comparing different aspects of tissue-organisation across species, we aim to uncover universal strategies and molecular mechanisms behind temperature resilience during gastrulation.
Novel technologies to facilitate discovery
To tackle biological complexity, we develop software and hardware tools, including machine learning algorithms for high-throughput imaging/analysis of organoids, methods to map mechanical properties in stem cells and lab automation solutions to improve experimental reproducibility.
Having uncovered key principles of early tissue self-organisation, we now look ahead to emerging frontiers. Advancing tissue engineering demands a holistic perspective—integrating geometry, biochemical cues, and cross-tissue connections with predictive modelling—to fully capture multicellular complexity. Our future aims are to:
1. Decipher how the interplay between global tissue geometry (shape, size, and dimension) and physical boundary conditions control patterning.
2. Develop novel methods to control tissue patterning using metabolites or hormones.
3.Incorporate complexity into developing tissues by including multiple cell types, such as lymphoid tissues and neuronal connections within developing tissues.
4. Apply theoretical and AI-based approaches to model mechanochemical interactions and predict tissue patterns.