Nassos Typas
Head of Molecular Systems Biology
athanasios.typas [at] embl.de
ORCID: 0000-0002-0797-9018
EditSystems microbiology

Head of Molecular Systems Biology
athanasios.typas [at] embl.de
ORCID: 0000-0002-0797-9018
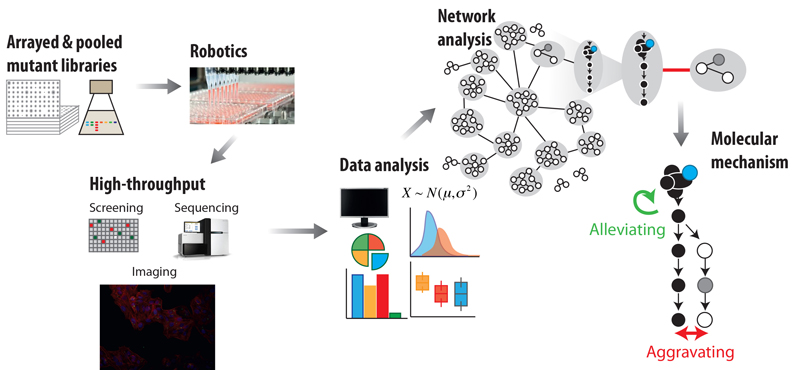
EditIn recent years, the explosion of sequencing efforts has provided a glimpse in the diversity of microbes that live on this planet, and the enormous functional potential they encode. We develop high-throughput quantitative approaches to shed light into gene function and organization in bacteria (figure 1). These approaches involve combining reverse genetics with quantitative genome-wide readouts to link genes to phenotypes and/or to known functional modules, such as pathways and protein complexes. We then use the data as starting points to generate mechanistic insights into targeted cellular processes, and to understand how function, regulation and cross-talk between cellular processes changes across evolution, impacting the phenotype.
Our main biological focus is on the bacterial envelope – its assembly, organization and ability to sense and transmit information. Working at the intersection between systems-based approaches and molecular mechanism, we have discovered key missing players of cell wall biosynthesis, established that conserved envelope machines are regulated and cross-talk in a rapidly evolving manner, and uncovered signalling pathways that the cell uses to monitor envelope defects. The cell envelope is how bacteria sense and interact with the environment, but also what protects them from external insults, such as antibiotics. This natural connection to antibiotics, has initiated to large line of research in the lab towards devising new strategies to target resistant pathogens and to solve current bottlenecks in antibiotic discovery. In the past, we have systematically profiled drug interactions in prominent Gram-negative and Gram-positive bacterial species, uncovering general principles that govern drug interactions and numerous synergies that are effective against clinical isolates of difficult to treat pathogens. We have also developed proteomics-based (together with the Savitski team) and chemical-genetics based ways to identify the mode-of-action of drugs, and their interaction at the resistance level (cross-resistance and collateral sensitivity).
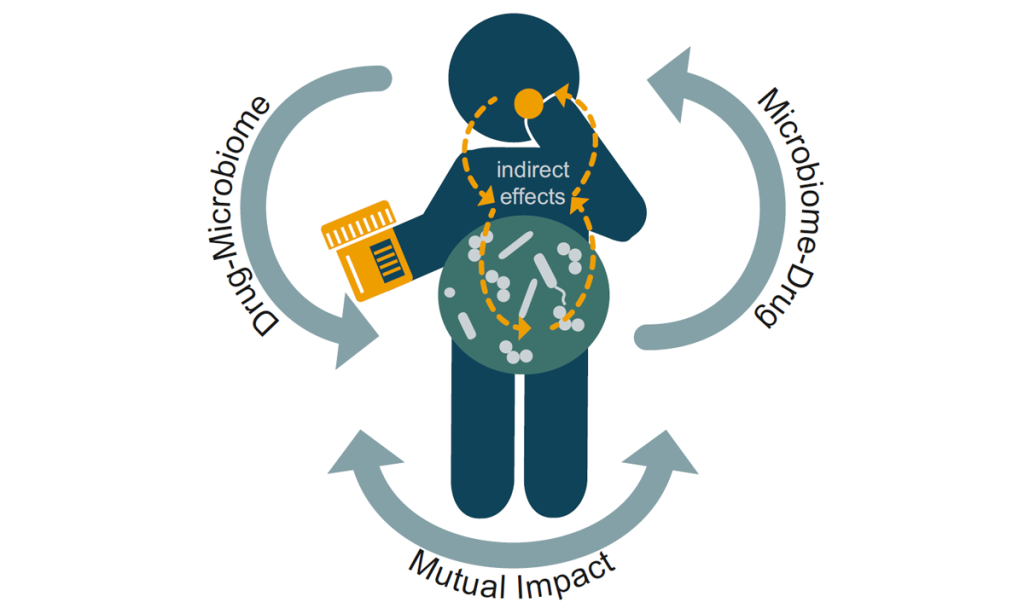
We have also recently moved our efforts to developing automated platforms and genome-wide approaches to study: a) the human gut microbiome and b) host-pathogen interactions. Over the past few years and together with the Bork, Patil, Zeller and more recently Zimmermann labs at EMBL, we have established systematic cultivation and phenotyping pipelines for the human gut microbiome. We have then used these in vitro microbiomics pipelines to study extensively the interactions of pharmaceuticals with the human gut microbiome (Figure 2), unravelling an unexpectedly high impact of non-antibiotic drugs on human gut microbes, a mechanism by which bacteria store drugs intracellularly, ways to mitigate the collateral damage of antibiotics to the gut microbiome, and emergent behaviours of communities to drug treatment. In the host-pathogen interface, we have used Salmonella as our model organism, to perform the first systematic host-pathogen protein-protein interaction study in the context of infection, map the host response during infection – unravelling a new role of cathepsins in cell death, and devise a labelling strategy to selectively and directly measure Salmonella’s metabolic fluxes during proliferation inside host cells.
We are broadly interested in the impact of mutations and horizontal gene transfer in the evolution of bacterial cellular networks. To tackle this, we are focusing on systems-based approaches in the E. coli pangenome and in related enterobacteria. On the drug front, we are interested in discovering novel ways to target pathogens, and in understanding and mitigating antibiotic resistance. Capitalizing on our unique in vitro “microbiomics” platforms, we aim to move some of our systems based approaches to abundant and key gut microbiota species, which still remain terra incognita. Together with 15 other labs at EMBL (and alumni) and as part of the Microbial Ecosystems Transversal Theme, we are working towards generating the tools, resources and functional information required for establishing from scratch new model species for the human gut microbiome. We are also expanding our experiment pipelines to be able to study the dynamics and properties of personalized microbiome communities, and aim to devise strategies for targeted strain replacement within them. This know-how, resources and automated pipelines will move in a new facility being established at EMBL on Microbial Automation and Culturomics.
In the host-pathogen front, our efforts have more towards understanding the tug of war that takes place between bacteria and phages. Building on our recent discovery of a new bacterial immune system and our know-how on host-pathogen interactions, we plan to use systematic genetic and biochemical approaches to understand the mechanisms, diversity and evolution of this complex and fascinating interface. In the long run, understanding bacterial immunity can open new doors for fighting bacterial pathogens by exploiting their internal weapons.