Jordi van Gestel
Group Leader
ORCID: 0000-0001-5598-5239
EditEvolution of microbial life cycles

Group Leader
ORCID: 0000-0001-5598-5239
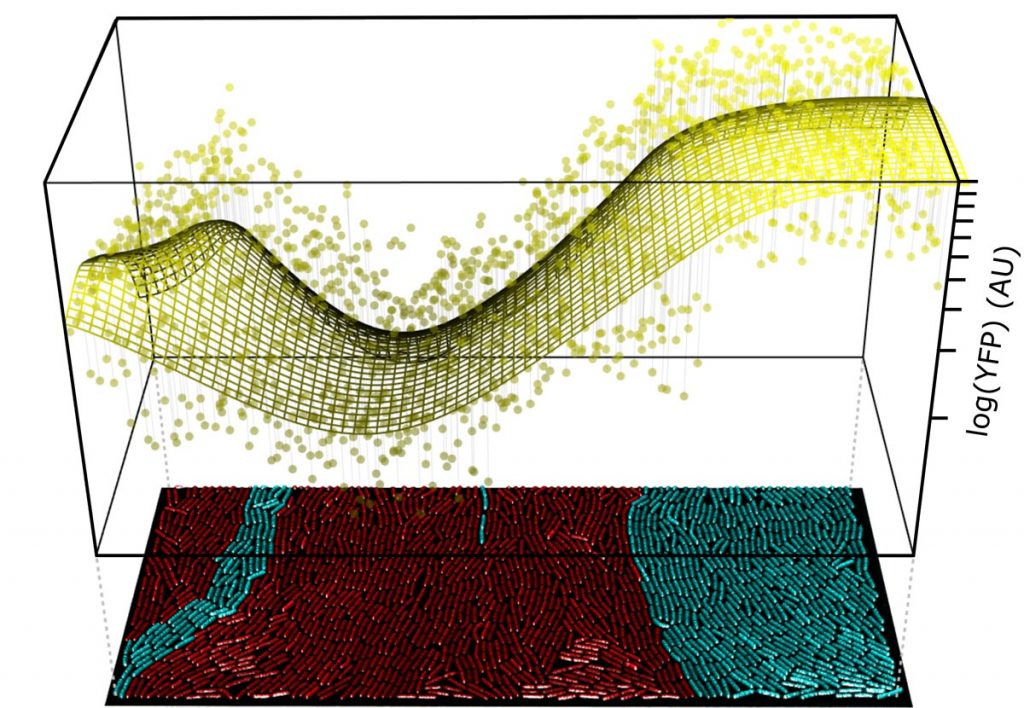
EditThe vast majority of both eukaryotic and prokaryotic biodiversity on our planet is microscopic. Although most microbes can propagate as single cells, many display primitive forms of collective development, such as cell-to-cell communication (i.e., quorum-sensing, Figure 1, van Gestel et al., Nat Comm 2021), cell differentiation (van Gestel et al., PLoS Biol 2015; van Gestel et al., Nature Eco & Evo 2020) and spatial patterning (van Gestel & Tarnita, PNAS 2017; van Gestel & Wagner, PLoS Biol 2021). Despite the widespread occurrence of such developmental phenotypes, their regulatory underpinnings and ecological implications often remain elusive. How do developmental mechanisms evolve in time, and how do they affect the emergent properties of collectives and their propagation in their natural ecology? We address these questions by studying how microbial development evolves under predation – one of the prominent ecological causes of evolutionary innovation.
Predator-prey interactions abound: in just a pinch of soil thousands of protozoan predators scavenge their bacterial prey (Figure 2). These predator-prey interactions unleash a fierce evolutionary arms race: bacteria evolve mechanisms that either prevent them from being phagocytized and/or support escape from the phagosome, while conversely protozoans evolve sophisticated hunting strategies to find, kill, and digest their prey. In this arms race, both predators and prey often form collectives that show adaptive emergent properties (Figure 3). For example, protozoans can invade bacterial communities through collective motility, while bacteria can withstand predation by adhering together. What is more, the tug of war between predator and prey can sometimes also profoundly change character, leading to the emergence of host-symbiont or host-pathogen interactions.
We study how predator-prey co-evolution impacts microbial development using an interdisciplinary approach – combining quantitative single-cell microscopy, microfluidics, functional genomics, high-content CRISPR libraries, experimental evolution, and mathematical modelling. Our goal is to identify the mechanisms that underlie predation and predation evasion, determine how these mechanisms are regulated and how their evolution impacts the interaction between protozoans and bacteria. Our starting point is the interaction between Bacillus subtilis and Dictyostelium discoideum, two soil-dwelling organisms with well-studied collective phenotypes, but, ultimately, we will explore natural communities of co-evolving protozoans and bacteria (Figure 3).
As part of both the Developmental Biology Unit and the Microbial Ecosystems Transversal Theme at EMBL, we have a growing network of collaborators within EMBL (e.g., Typas group, Vincent group) and outside (e.g., Gross lab at University of California San Francisco and Tarnita lab at Princeton University).
We are at the very beginning of uncovering the impact of predation on microbial development. Thus, many questions remain to be addressed, while others remain to be formulated. Some medium-term questions are: