2025
Planet Earth Award

Great news for the lab! You can check out the full EMBL article here.
Features in Nature Publishing Group


Some outreach about plankton, arts and science


Welcome to 2025 !

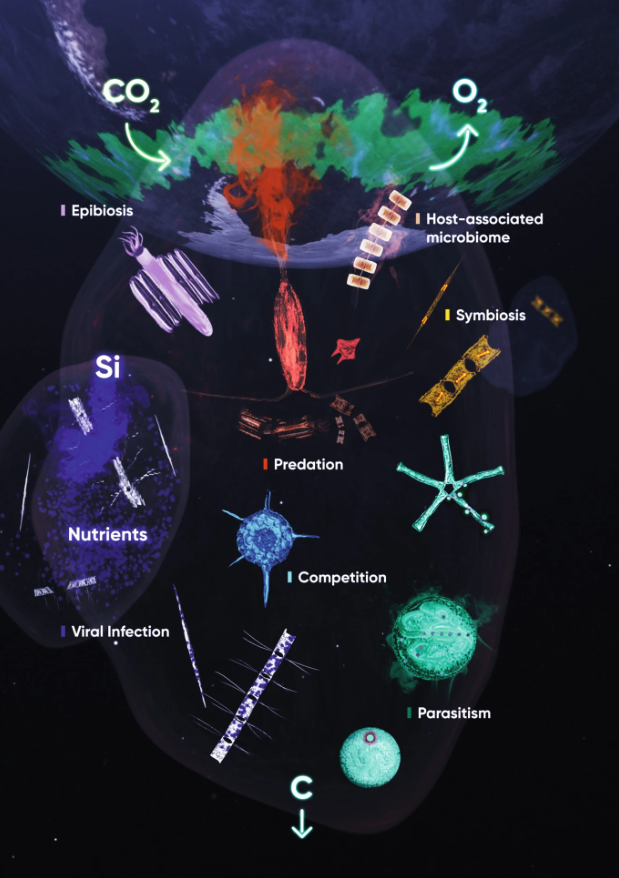
Symbiosis in marine unicellular eukaryotes

Great news for the lab! You can check out the full EMBL article here.





This marks the second year of the Vincent Lab ! Happy holidays and see you in 2025 !

Since September 2024 the lab counts two new members: Nina Guerin and Ellie Flaum. Nina and Ellie are two post docs that have both been awarded an EIPOD fellowship, in collaboration with Arnau Sebé-Pedrós (CRG) and Gautam Dey (EMBL) respectively. Get ready for some more Chaetoceros molecular and cellular biology.

Here are a few selected conferences where the lab has been in the past couple of months.





Since the kick-off of TREC in March 2023, our laboratory has been very active in the field, both on land and at sea. Soraya, Thomas, and Vanessa were lucky enough to embark on Tara, and Flora embarked for the fourth time as chief scientist between Bilbao and Cadix. We also spend a lot of time in the Advanced Mobile Lab, where Serena, Thomas, Kevin and Laurine have conducted many different types of experiments, and developed new approaches in marine microbiology. For us, the best part of the TREC expedition is when the Tara schooner meets up with the Advanced Mobile Lab, creating a unique opportunity to combine Tara’s holistic sampling strategy with targeted molecular biology using state-of-the-art equipment on the Advanced Mobile Lab.
Amir From, Gur Hevroni, Flora Vincent, Daniella Schatz, Carolina A. Martinez-Gutierrez, Frank O. Aylward and Assaf Vardi
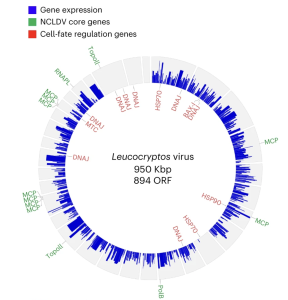
In this study we show how state-of-the-art single-cell approaches can be used to identify previously undescribed microbial interactions from complex marine samples, using viruses as an example.
Giant viruses (phylum Nucleocytoviricota) are globally distributed in aquatic ecosystems and play a fundamental role as evolutionary drivers of eukaryotic plankton. However, we lack knowledge of their native hosts, which limits our understanding of their life cycle and ecological importance. In 2018, we made a bet and decided to apply a single-cell RNA sequencing (scRNA-seq) approach to samples collected during a major mesocosm study, hoping it could help us discover new pairs of protist hosts and viruses. And it did!
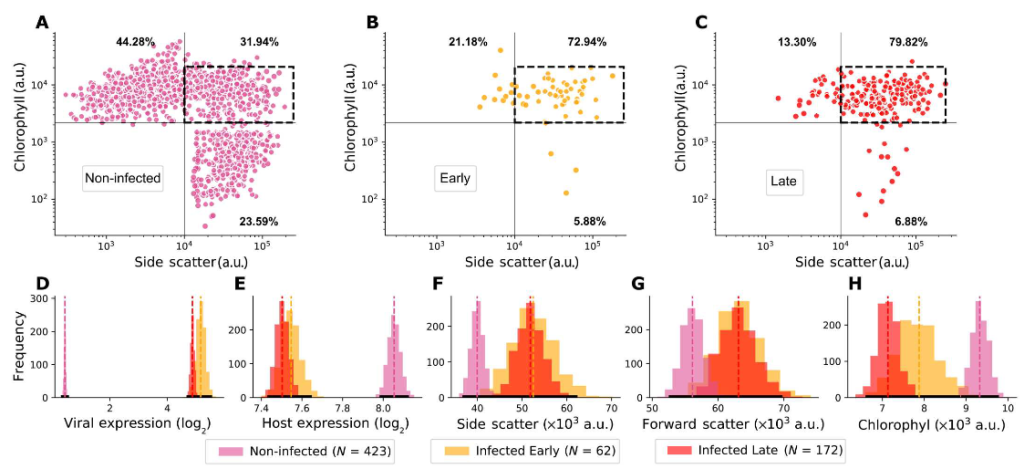
Gur Hevroni, Flora Vincent, Chuan Ku, Uri Sheyn, and Assaf Vardi
What are the dynamics of viral infection within natural blooms, and how does viral infection proceed at the single-cell level within complex communities? To address these questions, we applied single-cell RNA sequencing to profile the viral and host transcriptomes of 12,000 individual algal cells from an induced algal bloom. This involved collecting samples and sorting a target species based on FACS, in the shortest possible time frame. A consistent percentage of infected algae displayed the early phase of viral replication for several consecutive days, indicating a daily turnover and continuous virocell-associated metabolite production, potentially affecting the surrounding microbiome. These findings highlight the importance of studying the dynamics of microbial interactions in natural populations, at single-cell resolution, to better understand their impact on microbial food webs.
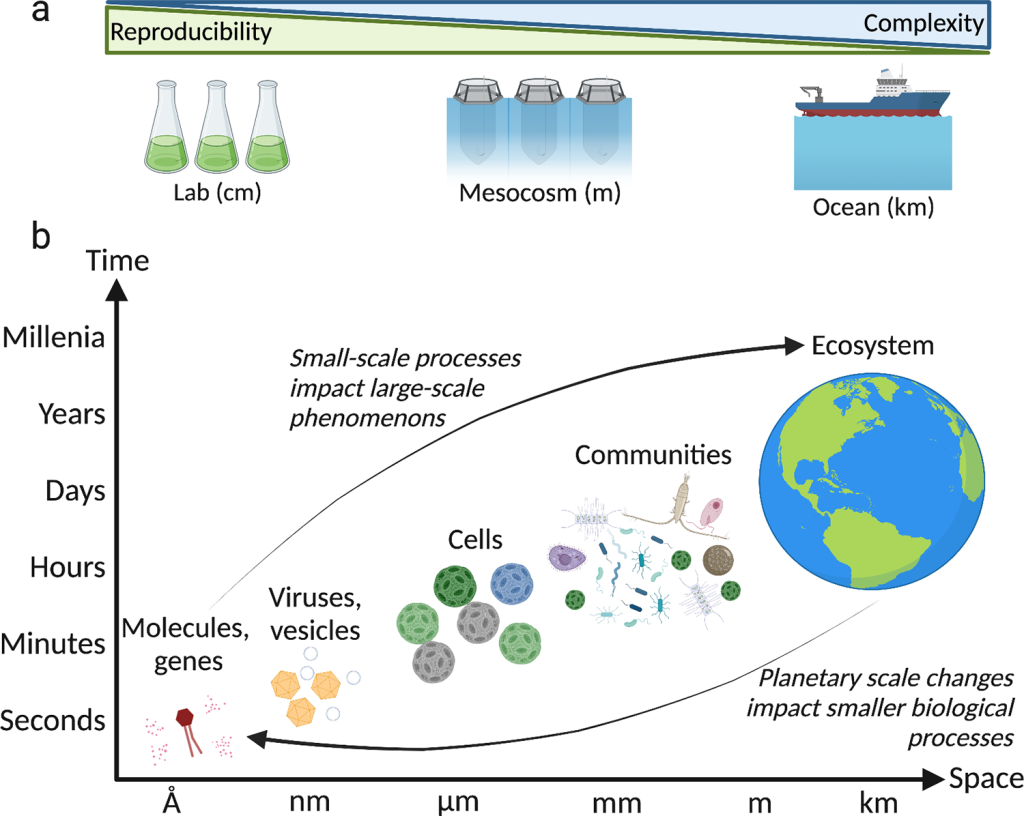
Flora Vincent, Assaf Vardi
One of the biggest challenges in marine microbiology is to assess the impact of microbial interactions across biological scales, from the single cell to the ecosystem level. Here, we argue that the advent of new methodologies and conceptual frameworks represents an exciting time to pursue these efforts and propose a set of important challenges for the field, taking aquatic viral infection as an example. Solutions include combining different scales of experimental setups, improving our ability to study non-cultivable interactions at the single-cell level, and establishing ecologically relevant and scalable model systems.
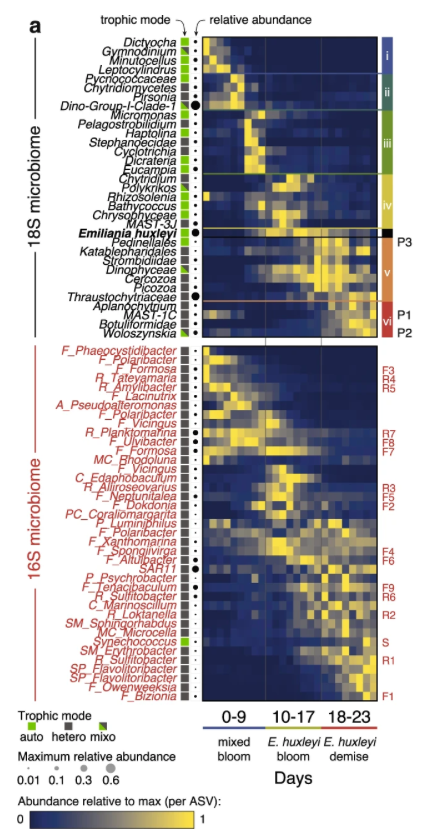
Flora Vincent, Matti Gralka, Guy Schleyer, Daniella Schatz, Miguel Cabrera-Brufau, Constanze Kuhlisch, Andreas Sichert, Silvia Vidal-Melgosa, Kyle Mayers, Noa Barak-Gavish, J. Michel Flores, Marta Masdeu-Navarro, Jorun Karin Egge, Aud Larsen, Jan-Hendrik Hehemann, Celia Marrasé, Rafel Simó, Otto X. Cordero & Assaf Vardi
We conducted a large-scale mesocosm experiment to quantify the impact of viruses on microbial communities and biogeochemical processes, during a major algal bloom, through a month-long time series combining single-cell assays, bulk biogeochemical measurements and microbial sequencing. By combining modelling and quantification of active viral infection at a single-cell resolution, we estimate that viral infection causes a 2–4 fold increase in per-cell rates of extracellular carbon release in the form of acidic polysaccharides and particulate inorganic carbon, two major contributors to carbon sinking in the deep ocean. This shows how microbial interactions happening at the molecular level can ultimately impact ecosystem processes related to carbon cycling, but also community composition. Co-first authors Flora and Matti wrote a “Behind The Paper” for it.
Flora Vincent & Chris Bowler
In this Chapter from ‘The Molecular Life of Diatoms” we highlight why diatoms, successful phytoplankton that play a major role in our ecosystem, are a great system to study the diversity and impact of microbial interactions across biological scales.
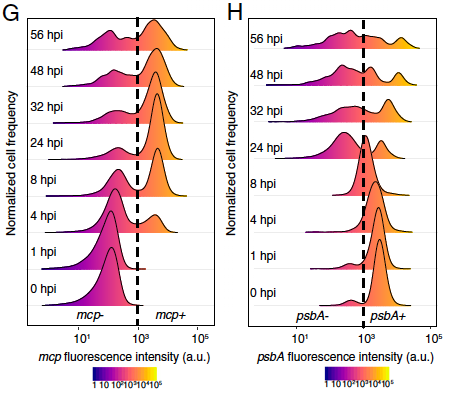
Flora Vincent, Uri Steyn, Ziv Porat, Daniella Schatz and Assan Vardi
Despite years of research in aquatic virology, we remain unable to estimate viral-induced mortality in the ocean and, consequently, to resolve viral impact on nutrient fluxes and microbial dynamics. Here, we assess active infection in algal single cells by subcellular visualization of virus and host transcripts using imaging flow cytometry coupled to mRNA FISH, revealing the coexistence of infected and noninfected subpopulations. We revisit major assumptions of a giant virus’ life cycle: cells can produce virions without lysing and can lyse without producing virions. In a natural algal bloom, only 25% of cells were infected, highlighting the importance of other mortality agents. Enrichment of infected cells in cell aggregates suggests potential host defence strategies. Our approach opens a mechanistic dimension to the study of microbial interactions, by developing single-cell ecology approaches in marine microbiology.
Flora J. Vincent, Sébastien Colin, Sarah Romac, Eleonora Scalco, Lucie Bittner, Yonara Garcia, Rubens M. Lopes, John R. Dolan, Adriana Zingone, Colomban de Vargas & Chris Bowler
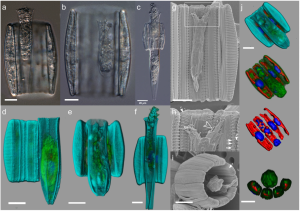
Diatoms are a diverse and ecologically important group of phytoplankton. Although most species are considered free living, several are known to interact with other organisms within the plankton. Here, we leverage the heterogeneous Tara Oceans dataset to describe a cosmopolitan and abundant symbiosis between diatom and ciliates, which combines high-content confocal microscopy, live imaging and single-cell sorting. As this symbiosis cannot be grown in the lab, this encourages us to push forward the ability to study microbial symbioses directly in nature.