
Matthias Wilmanns
Group Leader and Senior Scientist
ORCID: 0000-0002-4643-5435
EditStructure and function of molecular machinery for protein translocation across membranes

Group Leader and Senior Scientist
ORCID: 0000-0002-4643-5435
EditWe are investigating the overall structures of challenging protein complexes of biomedical relevance to address major research questions in the infection process of pathogenic bacteria but also in cancer, cardiology, and nephrology. To achieve these goals, we combine X-ray crystallography and single-particle cryo-electron microscopy and other complementary structural biology techniques. At the centre of our research interest is achieving an in-depth understanding of molecular translocation processes across membranes. A second research focus is to study how proteins react to external forces within the cell by means of molecular elasticity.
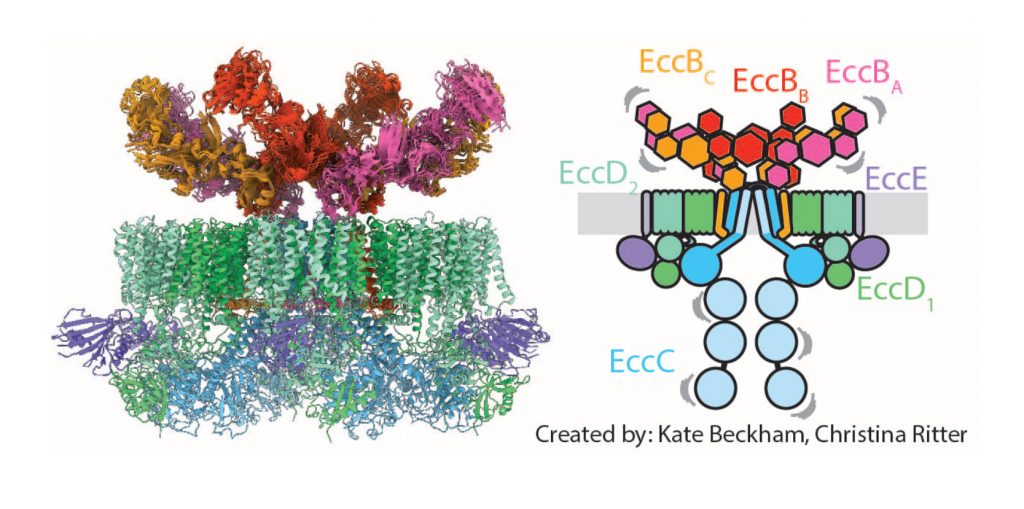
Pathogenic mycobacteria comprise distinct type VII secretion machineries that are unrelated to other bacterial secretion systems. These systems are key components for mycobacterial growth and infection. Using an integrative structural biology approach, our group has provided structural insight into the complete ESX-5 secretion apparatus. Following the initial low-resolution structure, we have now determined its high-resolution structure, revealing the substrate gating segment on the cytosolic side, the central pore that crosses the membrane, and a peculiar arrangement on the periplasmic side (Fig. 1). These findings provide ample opportunities to functionally study the overall secretion process.
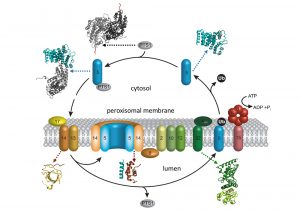
Peroxisomes are cell organelles that allow sequestered metabolic processes central to the viability of living organisms. As peroxisomes have no dedicated protein synthesis machinery, their function depends on the import of protein targets through a peroxisomal membrane translocon. Although the presence of such a translocon has been proven, to date any insight into the overall architecture is still missing. While our previous work has mainly focused on the structural elucidation of several translocon subcomplexes involved in substrate recognition and membrane docking (Fig. 2), our main future focus will be to unravel the structure of the entire translocon on its own.
Sarcomeres represent the fundamental unit in cardiac and skeletal muscle cells to provide a molecular framework for their basic function: contraction and relaxation. To cope with the substantial molecular forces generated by this process, sarcomeres are organised into complex networks of – in part – very long protein filament systems. Our past focus has been on three of those: titin, myomesin, and obscurin. By combining knowledge of their structures and single-molecule methods to measure their response to external forces, we have found reversible unfolding of exposed helical linkers to present a new novel mechanism of molecular protein elasticity. We are aiming to extend this knowledge into even larger filament complexes and to use functional methods to prove these mechanisms to be relevant under physiological conditions.
EMBL Hamburg c/o DESY, Notkestraße 85, 22607 Hamburg, Germany
Email: matthias.wilmanns [at] embl-hamburg.de