Kendrew and Philipson Awards Winners 2018
EMBL recognises the outstanding work of alumni with the John Kendrew and Lennart Philipson Awards
From Big Data to viral enzymes, EMBL alumni are tackling questions across the full range of science disciplines. This year’s alumni awards celebrate this diversity by showcasing the work of Nils Gehlenborg, winner of the 2018 John Kendrew Young Scientist Award, and Raffaele De Francesco, winner of the 2018 Lennart Philipson Award.
Big Data to find new questions

For centuries, data visualisation has helped humanity identify patterns and gain new insights. When London suffered a cholera outbreak in 1854, the English physician John Snow used a map to trace the source of the infection to the water pump in Broad Street, Soho. Now – in the era of Big Data – visualisation is more important than ever.
EMBL alumnus Nils Gehlenborg is pushing forward the visualisation of complex genomic and clinical datasets with highly innovative contributions. “I use computer science to build tools and visual interfaces that enable researchers to efficiently interact with biomedical data,” he explains.
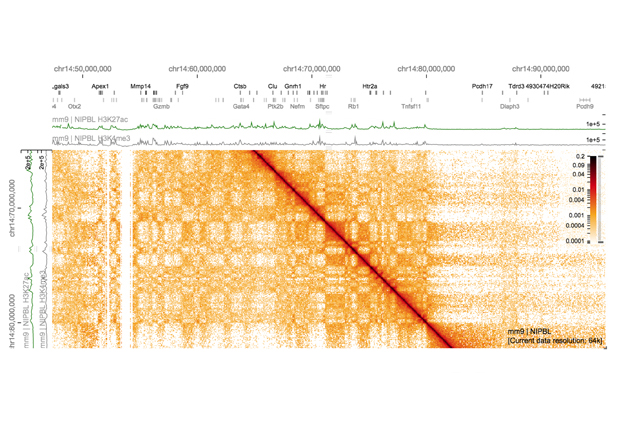
As a PhD student in the Brazma Group at EMBL-EBI, Gehlenborg worked on making large collections of gene expression data visually accessible so that biologists could discover patterns more easily. He is now Assistant Professor of Biomedical Informatics at Harvard Medical School, where he develops tools to visualise various types of data from large-scale cancer genomics studies such as The Cancer Genome Atlas. He also tackles visualisation problems across scales as co-investigator of the 4D Nucleome Data Coordination and Integration Center, funded by the US National Institutes of Health. With his team, he has created visualisation tools that allow scientists to see patterns at the chromosome level, and then to zoom down to find patterns at the level of DNA bases – the individual letters of the genetic sequence.
It’s a tool that helps us formulate new questions, rather than one which gives us all the answers
Gehlenborg’s lab is also exploring clinical applications of data visualisation by using data from electronic health records. “My long-term vision is that all the data generated by clinical analysis, sensors, and smartphones will be integrated,” he explains. “Rightly visualised, this will help doctors diagnose and treat patients using precision medicine. The visualisation and accessibility of the data will also help individuals understand how their health might be influenced by their behaviour.”
In the era of Big Data, biology is increasingly becoming a data-driven science, and this is changing the way we think about biological problems. “Traditionally, biology has been driven by hypotheses,” explains Gehlenborg. “You observe something in nature, come up with a hypothesis, and design an experiment to test it. But today we’re often collecting large amounts of data without a clear hypothesis. We try to find patterns and then maybe propose a hypothesis that we can test. This is where data visualisation is really strong. It’s a tool that helps us formulate new questions, rather than one which gives us all the answers.”
A medical revolution

EMBL alumnus Raffaele De Francesco’s work on the Hepatitis C virus (HCV) led to a medical revolution. Previously, certain chronic viral infections had been kept under control using drugs, but it had never been possible to cure them completely by removing the virus from the body. De Francesco’s work allowed Hepatitis C to become the first case in the history of medicine in which a chronic viral infection was cured with direct antiviral agents.
I wanted to find the virus’ Achilles’ heel and hit it with antiviral agents
Although the genome of HCV had been isolated and sequenced, finding a cure for Hepatitis C still presented a challenge. Liver cells are difficult to grow in cell culture, and the virus didn’t function as well in cultured cells as it does in the human body. It took more than a decade to find the right conditions to grow the virus in the laboratory. It was then necessary to develop biochemical tools and assays that would help researchers find ways of interfering with the virus’ enzymes – preventing it from functioning.

These technical challenges, as well as the huge medical need, pushed De Francesco to work with all the molecular biology and biochemistry tools available, to understand what each part of the virus’ genome does. “I wanted to find the virus’ Achilles’ heel and hit it with antiviral agents,” he explains.
In his lab at the Institute for Research in Molecular Biology (IRBM) in Pomezia, Italy, he identified, purified and developed in vitro assays for two viral key proteins, NS3/4A protease and NS5B polymerase. His discoveries allowed the scientific community to start screening for agents to inhibit the two viral enzymes, eventually leading to a cure for Hepatitis C.
My work in Cortese’s group was really pivotal in starting my career as an independent scientist
Now Head of Virology at INGM, the National Institute for Molecular Genetics, in Milan, Italy, De Francesco is clear about what he gained from his time in Riccardo Cortese‘s group at EMBL. “Riccardo gave me a lot of guidance but at the same time enough autonomy and freedom to follow my own interests,” he explains. “My work in his group was really pivotal in starting my career as an independent scientist.”