First interactive model of human cell division
Real-time tracking of proteins during mitosis is now possible using a 4D computer model made by EMBL scientists
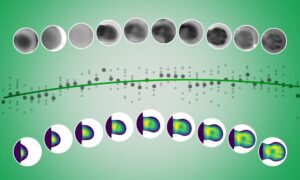
Mitosis – how one cell divides and becomes two – is one of the fundamental processes of life. Researchers at EMBL have now produced the first interactive map of proteins that make our cells divide, allowing users to track exactly where and in which groups the proteins drive the division process forward. This first dynamic protein atlas of human cell division is published in Nature today.
In 2010, a large study led by the same EMBL group identified which parts of the human genome are required for a human cell to divide, as part of the EU MitoCheck project. But cells don’t run on genomic DNA; they run on the proteins it encodes. Proteins carry out most of the work in a cell, forming the cell’s operational level. Processes like mitosis require the tight coordination of hundreds of different proteins in space and time. Proteins often work in groups, similar to the specialist teams of construction workers on a large building site.
“Until now, individual labs have mostly been looking at single proteins in living cells,” says Jan Ellenberg, the group leader at EMBL who led the project. “Supported by the follow-up EU project MitoSys we were now able to take a systems approach, and look at the bigger picture by studying the dynamic networks many proteins form in living human cells.”
The resulting Mitotic Cell Atlas integrates these data in an interactive 4D computer model. In this public resource, scientists can freely choose any combination of mitotic proteins and see in real time where and with whom they work during cell division.
Five different proteins are tracked during cell division (from metaphase to telophase): AURKB (red), NUP107 (green), CENPA (purple), CEP192 (yellow), and TUBB4B (cyan). The video represents what users could create by themselves when using the mitotic cell atlas homepage: www.mitocheck.org/mitotic_cell_atlas/
Sharing the tools to make more cell atlases
Cell division is an essential process of life. When it goes wrong, defects like fertility issues and cancer can occur. Ellenberg: “Besides mitosis, the technologies developed here can be used to study proteins that drive other cellular functions, for example cell death, cell migration or metastasis of cancer cells. By looking at the dynamic networks these proteins form, we can identify critical vulnerabilities, points where there’s only one protein responsible to link two tasks together without a back-up.”
Looking at disease relevant processes from a dynamic network point of view provides a new perspective to find their critical links, where they can be cut or rewired to strengthen them. To enable more such studies in the future, the experimental methods, the quantitative microscopy platform, and the code to create dynamic protein atlases are now openly available for others to use.
Counting proteins in living cells
The current study looked at HeLa cells, a widely used line of human cancer cells. 28 proteins that are important for mitosis were made fluorescent mostly by CRISPR/Cas genome editing. These proteins were then tracked using 3D confocal microscopy, to see where in the cell they’re located at each point in time. The microscope is so sensitive that it’s even possible to count the proteins, so researchers know now if there are 100, 1000 or 10.000 proteins in a certain location. For all proteins, these data were integrated into an interactive computer model – the creation of which was actually the largest part of the project.
In total, there are about 600 different proteins involved in mitosis in human cells. Completing the dataset for all 600 would allow scientists to fully understand the transmission of information within a dividing cell, and how decisions – like going from one cell cycle phase to the next – are made. This will take several more years of work. “At EMBL, we’re constantly adding information to the atlas by imaging more proteins in the same standardized way” says Stephanie Alexander, research manager in EMBL’s Ellenberg group. “In the long run, a full overview of all the cell’s proteins will allow us to see how different important processes of life, like cell division and cell death for example, are linked to one another. You can only understand this from a network point of view.”
The EMBL team collaborated with researchers from the Research Institute of Molecular Pathology (IMP) in Vienna, Austria. The work was supported by grants from the EU-FP7-MitoSys, EU-FP7-SystemsMicroscopy NoE, EU-H2020-iNEXT, as well as by the European Molecular Biology Laboratory.