Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis
Nature Biotechnology 6 September 2021
10.1038/s41587-021-01006-2
Researchers have combined spatial gene expression information with single-cell genomics data to create a high-resolution atlas of mouse organogenesis
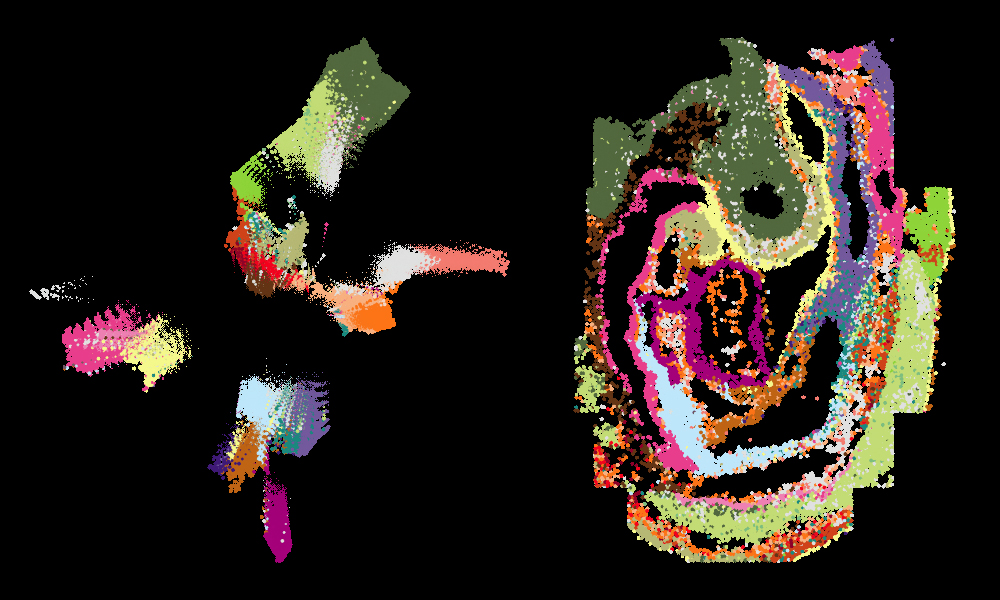
High-resolution gene expression maps have been combined with single-cell genomics data to create a new resource for studying how cells adopt different identities during mammalian development. The Spatial Mouse Atlas is the result of a collaboration between researchers at EMBL’s European Bioinformatics Institute (EMBL-EBI), the Babraham Institute, the Cambridge Stem Cell Institute, the Cancer Research UK (CRUK) Cambridge Institute, and the California Institute of Technology (Caltech) and colleagues.
Cell fate decisions determine how cells develop into different cell types. The development of different cell types eventually leads to the formation of all the different tissues in the body. This complex process involves many different signals from surrounding tissues, as well as mechanical constraints, epigenetic modifications and changes to gene expression. These factors create unique cell and tissue types, which eventually give rise to all major organs in a process called organogenesis.
Studies using single-cell RNA sequencing (scRNAseq) have provided insights into how the molecular landscape of cells in the mouse embryo changes during early development. However, scRNAseq methods require the cells in the embryo to be dissociated – or separated – which means spatial information is lost.
This study, published in Nature Biotechnology, combines scRNAseq data with spatially-resolved expression profiles to generate an atlas of gene expression at single-cell resolution across the entire embryo.
“Methodologically, I think this is one of the most exciting examples of integrating spatially resolved transcriptomics and single-cell sequencing to create a new resource for the scientific community,” said John Marioni, Head of Research at EMBL-EBI. “It’s also work we can build upon to include more target genes across different stages of development.”
The researchers carried out this study using 8–12 somite stage mouse embryos. This stage of development was of particular interest because it is when the cells within the embryo start to differentiate, or become a specific cell type. The researchers applied an image-based single-cell transcriptomics method – seqFISH (developed by collaborators at Caltech) – to detect 387 target genes. They combined this information with scRNAseq data to produce an atlas of cell types found across this stage of mouse development.
“For this study we needed to process, analyse and integrate a lot of data sets to enable us to map the identities of cells onto a spatial reference,” said Shila Ghazanfar, Research Associate at CRUK Cambridge Institute. “This new approach means that we can take a section of the embryo and essentially paint on the cell types using different colours to place cell type context on the anatomy of the embryo at single-cell resolution.”
Tim Lohoff, a PhD student at the Babraham Institute and lead author on the paper, commented: “Previously, researchers have been able to measure gene expression profiles for embryonic cells. However, it has long been a technological challenge to link these profiles to their original spatial location. Our work has been able to overcome this challenge, providing critical information about how the spatial environments of cells are critical to organ development, and opening up new avenues of research.”
The researchers were able to use the Spatial Mouse Atlas they created to uncover new insights into mouse development. They were able to find previously unreported gene expression information within the developing brain and gut tube of the mouse. This new approach provides a robust framework for future studies looking at spatial gene expression, both in the mouse and potentially other biological systems.
“This landmark study has already allowed us to pinpoint some of the molecular events that lead to the development of organs in mice. By providing a more detailed blueprint for development shared between mammals, this study could help to further advance the possibility of generating cell types in a dish for regenerative medicine.” said Professor Wolf Reik, Group leader in the Epigenetics research programme at the Babraham Institute.
Nature Biotechnology 6 September 2021
10.1038/s41587-021-01006-2