Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions
Nature Microbiology 24 December 2021
10.1038/s41564-021-01011-w
Researchers have identified hundreds of new bacterial species and viruses in the human skin microbiome
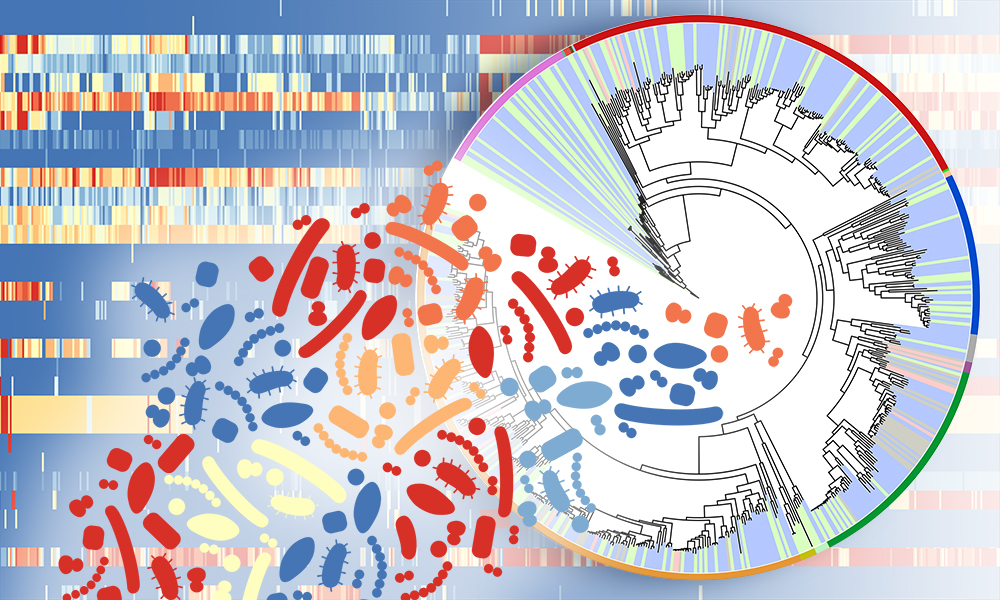
Researchers at EMBL’s European Bioinformatics Institute (EMBL-EBI), the National Institutes of Health (NIH) National Human Genome Research Institute (NHGRI), the NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases, and colleagues have identified new bacterial and fungal species, as well as viruses in the human skin microbiome.
Microbiomes – communities of microorganisms – are found everywhere, from the oceans and soil to the human gut and the surface of the skin. The skin microbiome is thought to play a key role in skin health and disease. Certain microorganisms in the skin microbiome are associated with different skin conditions, including acne and eczema.
In this study, published in Nature Microbiology, the researchers sequenced the genomes of the microorganisms detected within 594 samples taken from various skin surfaces of 12 healthy volunteers. Combining traditional laboratory cultivation with a metagenomic sequencing approach, the researchers were able to create the Skin Microbial Genome Collection (SMGC) – a collection of reference genomes for the human skin microbiome.
“We discovered thousands of viral sequences including many jumbo phages – very big viruses that infect bacteria – most commonly on the surface of the hands and feet of our volunteers,” said Sara Kashaf, PhD student at the NIH and EMBL-EBI. “These areas of the body have highly diverse microbiomes, which makes sense because we’re constantly using our hands to touch new things in our environment. Our future work will aim to understand what these different microbes are doing within these communities.”
The researchers used extensive lab culturing methods, metagenomic sequencing, and publicly-available skin metagenomes to gain these new insights into the extent of diversity within the skin microbiome. This study primarily focused on individuals from North America, but future work will aim to expand the SMGC using samples from different populations.
“This work is a major step toward obtaining a complete genomic blueprint of the skin microbiome,” said Julie Segre, Senior Investigator at the NHGRI. “We hope these data will support future investigations improving our understanding of skin health and disease.”
The SMGC collection includes 174 previously unknown bacterial species, four new eukaryotes and 20 new jumbo phages – viruses with a genome larger than 200 kilobases, 3–5 times larger than an average virus. Researchers have previously been working with an incomplete knowledge of the bacterial composition of the skin microbiome. However, this research expands the catalogue of known skin bacteria by 26%.
“In addition to the bacteria and viruses that we normally recover in metagenomics, we also found twelve genomes from single celled eukaryotes – fungi, like yeast – from the human skin. Some of these genomes were already known, such as Malassezia globosa, which has been associated with the healthy skin mycobiome but has also been associated with conditions such as dandruff. Using the same methods that recovered eight known genomes gives us confidence in the four novel eukaryotes that we found,” says Rob Finn, Group Leader at EMBL-EBI. “Looking at the patterns of these novel eukaryotes revealed that one of them was very common across the volunteers, and may be found on many of us.”
All of these data will be made freely accessible soon as a new genome catalog in the MGnify data resource, where researchers can find a huge range of microbiome datasets including for the human gut.
Nature Microbiology 24 December 2021
10.1038/s41564-021-01011-w