MASSIFly efficient
In September 2015, MASSIF-1 processed its 10,000th crystal, less than one year after the beamline became operational, proving its remarkable efficiency and importance to the scientific community.
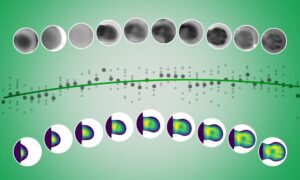
MASSIF-1, the first beamline of the Massively Automated Sample Selection Integrated Facility, is jointly operated by ESRF and EMBL, in Grenoble. This ground-breaking tool provides European structural biologists with a fully automated service for sample evaluation and data collection from crystals of macromolecules. Scientists feed their experimental requirements into the beamline software while their samples enter a queuing system. The crystals are then located and screened automatically, and the data acquired without any human intervention.
“The world-leading automation developed on MASSIF-1 means that as much data as possible is extracted from every sample and the robot never gets tired,” explains Matthew Bowler, MASSIF-1 beamline scientist at EMBL Grenoble.
This system both frees up the time of the scientists – who can focus better on analyzing the collected data – and improves the quality and the quantity of information collected. The automatic routine is better at locating the crystals and choosing the right angle for the beam than the human eye. In addition, more crystals can be efficiently screened: this aspect was crucial for the 10,000th crystal, the NtrX protein from Brucella abortus.
The world-leading automation developed on MASSIF-1 means that as much data as possible is extracted from every sample and the robot never gets tired
NtrX is made of 3 well-characterised domains, but their 3D arrangement has never been described. To tackle this question Ignacio Fernandez, PhD student from the Fundacion Instituto Leloir, Buenos Aires, Argentina, visiting EMBL Grenoble as one of six young Argentinean scientists supported by the Argentinian Ministry of Science, Technology and Productive Innovation, and Irina Cornaciu, a postdoctoral fellow at EMBL Grenoble, produced and crystalised the protein in different states: active, inactive, with its target nucleotide attached, without its target nucleotide… In total they screened through 10,000 different conditions to generate over 100 crystals.
“To my knowledge, MASSIF-1 is the only beamline that can cope with the volume of crystals we generated, and screen them rapidly,” explains Josan Márquez group leader at EMBL Grenoble. “It was crucial to the success of this project.” More generally, describing the 3D organisation of large macromolecules is an increasing, but challenging, necessity for structural biologists; such analyses require the ability to consistently screen large numbers of crystals, which would be very time-consuming and unreliable with more traditional human-driven techniques, making MASSIF a necessary improvement. However, interpretation of the 3D arrangement of the three domains of NtrX can only be performed by humans, so the final results will come later.