Enzyme with a dual-purpose loop
Unexpected results: structure of Death Associated Protein Kinases (DAPK) reveals a dual-purpose loop
“I really thought I had done something wrong!” says Bertrand Simon who has just completed his PhD in the group of Matthias Wilmanns at EMBL in Hamburg. Simon was studying a group of enzymes known as Death Associated Protein Kinases (DAPK), trying to understand how their activity is regulated. These enzymes can trigger the death of cells, ensuring they die off if they get out of check.
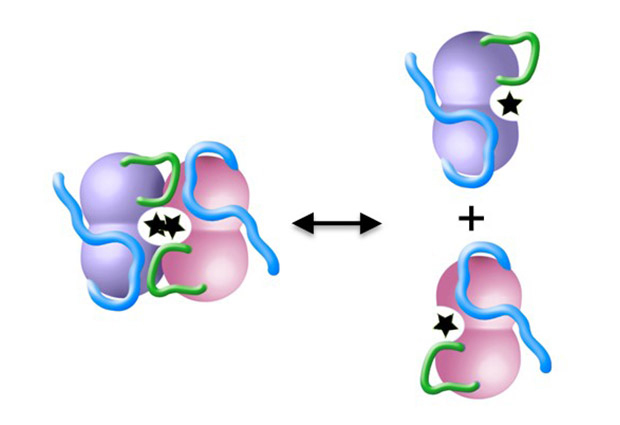
Simon was taking a closer look at the 3D molecular structure of several DAPK molecules. “DAPK has an inactive form and an active form” explains Simon, “We wanted to show exactly how two active DAPK molecules – each known as a ‘monomer’ – bind together to make up the inactive ‘dimer’ form of the enzyme.”
In the loop
The 3D structure of a protein arises when a string of amino acids folds together – much like a length of string can be scrunched together to form a ball in the palm of your hand. Although the string of amino acids is folded in and around itself, some parts of the string will still be exposed and can interact with other molecules.
“Looking at the structure, we identified a small loop of amino acids that we thought could be important for binding two DAPK molecules together,” explains Simon, who set about collecting evidence to support this assumption. “This looked like a small and straightforward project that would have finished off my PhD thesis nicely and added important information to our understanding of how this enzyme is regulated,” he states. But as so often in science, things didn’t turn out quite as expected.
This looked like a small and straightforward project that would have finished off my PhD thesis nicely
Several 3D molecular structures of DAPK had already been solved by previous group members and collaborators Andrew McCarthy and Darren Hart from EMBL in Grenoble using structural biology methods, and deposited in the Protein Data Bank Europe (PDBe). Using these structures, Simon was able to identify and then systematically mutate the amino acids found at and around the loop he was interested in, and so show exactly which parts are significant for molecule binding.
New hypothesis
One by one Simon exchanged the arginine amino acids along and around the loop for a reverse-charged amino acid, glutamate. The project started off nicely with everything working as planned. “All my experiments were going really well – something I had never experienced before!” says Simon with a smile. “The experiments showed that the monomers couldn’t bind together to form dimers without this little loop intact,” he adds.
Encouraged by this, and validated by additional structural biology experiments carried out by the Small Angle X-ray Scattering group of Dmitri Svergun in Hamburg, Simon introduced the mutated proteins into the cell environment to show that the effect was seen here too. “A change in enzyme activity after we had mutated a part of the protein would indicate we’d hit the right spot,” explains Simon. And nothing. “The protein was dead! Zero activity. It wasn’t even binding to its normal signaling partner, an activity regulator protein called calmodulin, which had been carefully prepared the Schultz group at EMBL in Heidelberg,” he says. “I thought, oops, I must have done something wrong!”
In a paper published in Science Signaling in 2010, Wilmanns and colleagues had already presented the 3D structure of a DAPK monomer bound to calmodulin. “The binding site for calmodulin was somewhere completely different to this small loop I was manipulating – it just didn’t make any sense that the calmodulin binding wasn’t working anymore,” he says.
There was nothing in the scientific literature that might explain what we were seeing
Simon repeated the experiments again, and again and again. “After about two months with no change in result, I decided I needed to start thinking in a different direction,” he says. Simon developed a new hypothesis: “I considered the results I had been collecting as statements, and built a hypothesis around them and other information we knew from previous work done on DAPK in our group.”
Simon presented his new hypothesis to his supervisor Wilmanns. “There was nothing in the scientific literature that might explain what we were seeing,” explains Wilmanns, “but the effects were so strong it was clear something interesting was going on.”
Dual purpose
The work published in Structure, suggests that the small loop is crucial for both dimer formation and calmodulin binding. A renewed look at the structure published in 2010 shows that the small loop plays a much more significant role there too, as Simon hypothesizes. “When we published the structure in 2010, we assumed the loop we were seeing was just an artefact of the sample preparation process,” says Wilmanns. “Now we think this small loop is needed to recruit calmodulin to DAPK in preparation for binding, hence without it, calmodulin binding does not work,” he adds.

“The new hypothesis is quite a complex idea, but it is the only logical explanation that brings all the results together,” says Simon.
“This is very satisfying and exciting,” concludes Wilmanns, “what started as a small side project, unearthed a complex and important signaling pathway within this group of kinases. It goes to show, you can’t always plan science!”