Subtleties in Clathrin heavy chain binding boxes provide selectivity among adaptor proteins of budding yeast
Nature Communications 7 November 2024
10.1038/s41467-024-54037-z
Key insights into a mechanism via which cells bring in cargo from outside
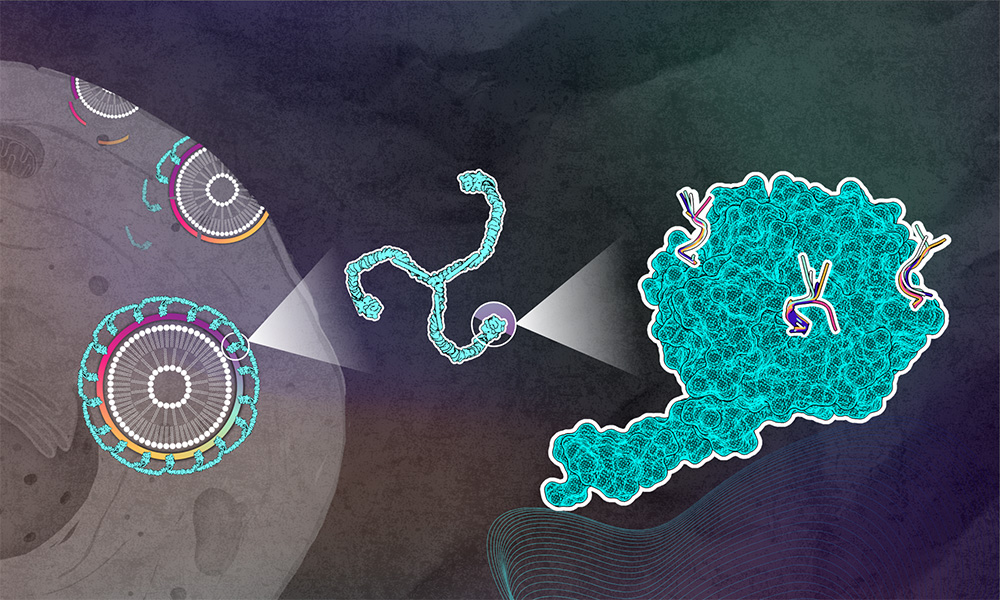
The cellular membrane controls the passage of substances in and out of the cell. Some larger cargo, such as membrane proteins, nutrients, and pathogens, enter the cell via a process known as clathrin-mediated endocytosis.
To gain a deeper understanding of this important cellular process, the researchers in the García Alai Team at EMBL Hamburg and CSSB in collaboration with the Uetrecht Group employed a combination of integrative biophysical and structural approaches together with in vivo functional experiments to take a closer look at the interactions between the protein clathrin and various adaptor proteins in budding yeast cells.
During clathrin-mediated endocytosis, adaptor proteins bind to the cell membrane and to the protein clathrin, assisting in engulfing the cargo in a clathrin-coated vesicle.
“The cargo is essentially sucked into the cell during clathrin-mediated endocytosis. The process is actually quite speedy; everything takes between 60 and 120 seconds – from the recruitment of the early adaptor proteins to the membrane to the excision of a vesicle,” explained Lucas Defelipe, postdoc in the García Alai Team and the study’s first author.
The clathrin protein has a three-legged shape and forms cages of different sizes to coat vesicles. At the end of each ‘clathrin leg’, there is the N-terminal domain (NTD) that interacts with adaptor proteins, linking this scaffold to the cellular membrane. Each NTD has three to four adaptor protein binding sites known as boxes.
To understand the role of these boxes in clathrin-mediated endocytois, the researchers needed to find out whether these NTD boxes have target-specific binding capabilities.
The clathrin heavy chain’s N-terminal domain bound to peptides from specific adaptor proteins was examined using X-ray crystallography at EMBL Hamburg beamlines P13 and P14. “All our structures show that each peptide binds to three different binding pockets known as Clathrin box, Arrestin box, and W-box,” explained María García-Alai, EMBL Team Leader and the study’s corresponding author. “In fact, all the peptides were able to adapt to fit into each of these boxes by adopting different conformations.”
Next, the researchers wanted to understand whether the adaptor proteins have any preference for the different boxes. Are they competing with each other to bind to a specific box? To understand the selectivity of adaptors for clathrin, the group conducted several native mass spectrometry (MS) competition experiments at EMBL’s Sample Preparation and Characterisation Facility. “These experiments revealed that adaptor protein Ent5 is the strongest binder to all NTD boxes, and more importantly, showed that the adaptor proteins Ent1 and Ent2 do in fact have binding preferences,” noted CSSB Group Leader Charlotte Uetrecht, whose group specialises in conducting native MS experiments.
To observe Ent1 and Ent2 preferences in the cellular context, the García-Alia Team performed experiments at CSSB’s Advanced Light and Fluorescence Microscopy (ALFM) Facility. As clathrin-mediated endocytosis is such a fast process, the scientists used an inhibitor to slow down the proteins. They tagged Ent1 and Ent2 with fluorescent proteins and then conducted FRET experiments, which report on the close spatial proximity of proteins, to observe their interactions with clathrin’s NTD boxes. “Our results indicate a preferential binding of Ent1 to the Clathrin box and of Ent2 to the Arrestin box,” noted ALFM Facility Head Roland Thuenauer. “We also noticed that Ent1 demonstrates a stronger interaction with clathrin.”
“As Ent1 and Ent2 were previously thought to be functionally redundant, these results are very exciting,” stated Defelipe. “Ent1’s stronger bond with clathrin also helps explain its functional divergence towards actin binding. Overall, we have shown that adaptor protein selectivity and its competitive binding of clathrin results in functional specialisation.”
This article has been adapted from a news article by the Centre for Structural Systems Biology (CSSB). Find the original story at the CSSB website here.
Nature Communications 7 November 2024
10.1038/s41467-024-54037-z