
“Structurally” sound
Connecting structure to function in five decades of research
By Julia Mahamid, Kristina Djinovic-Carugo, Janet Thornton
Structural biology investigates the three-dimensional structure of biological molecules, such as proteins and nucleic acids, and tries to understand their function through this lens. The journey of structural biology in the last five decades has been one of continuous technological progress coupled with exciting biological insights, greatly expanding our understanding of how life functions at the molecular level.
During this time, EMBL has been a world leader in the field, through innovative technological and methodological developments, novel biological discoveries, hosting of databases used by millions of scientists worldwide, and the provision of state-of-the-art structural biology services.
Crystallography advances
In 1975, just a year after EMBL was created, its founders, some of whom were pioneering structural biologists, took the decision to establish the next two sites (then called ‘outstations’) in Hamburg and Grenoble, on campuses with sources of high-energy beams/particles that could be used for structural biology experiments. EMBL Hamburg is situated on the campus of the German Synchrotron Research Centre (DESY), while EMBL Grenoble shares the EPN Campus with the European Synchrotron Radiation Facility (ESRF), the Institut Laue Langevin (ILL), and the French national Institut de Biologie Structurale (IBS).
Both EMBL Hamburg and Grenoble provide a rich environment for instrumentation engineers, scientists, and service specialists to collaborate and innovate, advancing structural biology methods as well as their use in understanding biological processes.
Many of these developments have focused on improving and automating techniques related to macromolecular crystallography. Researchers studying the three-dimensional structure of a macromolecule (e.g. a protein) need to first isolate it in its purest form from a biological sample that has thousands of other macromolecules. The isolated protein is then induced to form crystals – an orderly arrangement of molecules – whose atomic structure can then be determined by analysing the way they scatter X-rays or other energy beams.
In the early days of structural biology, methods for isolating, harvesting, and analysing protein crystals, as well as for interpreting the data obtained afterwards were manual, laborious, and time-consuming. Over the past five decades, however, EMBL researchers and engineers have introduced an unprecedented degree of automation in macromolecular crystallography, in addition to pioneering new techniques, thus revolutionising both crystallisation procedures and the use of beamlines.
For example, in 1990, EMBL Hamburg produced the first online imaging plate scanner for protein crystallography, which has been commercialised and widely used. EMBL Hamburg has also been leading the way in a technique called small-angle X-ray scattering (SAXS), which was used, for example, by EMBL scientists in collaboration with BioNTech, Johannes Gutenberg University Mainz, and other partners, to study how mRNA can be better packaged and delivered into human cells. This crucial research supported the mRNA nanomedicine technologies that were necessary for ensuring the quality and efficiency of mRNA drugs and vaccines, such as those developed during the COVID-19 pandemic. It is also worth noting that SAXS, by virtue of not requiring crystallisation, allows us to study molecules in solution, perhaps a more natural state for proteins than crystals.

EMBL Grenoble has a decennial track record in innovative technological development with over 100 in-house developed instruments deployed at 26 synchrotrons around the world. These developments included CrystalDirect™, an automated protein harvester, which EMBL Grenoble researchers integrated with the fully automated beamline MASSIF-1. This unique combination of structural biology technologies allows some of the key steps in protein crystallography to be completely automated. This service is now available to scientific users worldwide for use in their projects.
Partnerships and collaborations have played a key role in such advancements. In 2002, EMBL, ESRF, ILL, and IBS signed a memorandum of understanding (MoU) to create the Partnership for Structural Biology (PSB) on the European Photon and Neutron (EPN) science campus in Grenoble, following in the footsteps of previous MoUs between EMBL and ESRF in 1992 and 1997. The PSB has played a key role in improving the ease of access to advanced structural biology infrastructure for researchers in Europe as well as globally.

Cryo-electron microscopy and tomography – from large and flexible complexes to seeing deeper inside cells
While macromolecular crystallography allows researchers to determine the structure of an isolated protein with a high degree of precision, researchers have also been perfecting techniques that allow us to ‘see’ large and flexible macromolecular complexes using advanced electron microscopy. In the early 1980s, EMBL Heidelberg Group Leader Jacques Dubochet and technician Alasdair McDowall discovered a method to rapidly cool biomolecules in solution in such a way that their structure could be preserved – a process called vitrification.
This critical advance laid the groundwork for the rise of cryogenic electron microscopy (cryo-EM), a method which allows complex molecular machines to be imaged accurately. Dubochet would go on to win the Nobel Prize in Chemistry in 2017 for this work. Over the last decade, cryo-EM has become the method of choice for complex macromolecular structure determination, owing to the “resolution revolution” – a term coined by EMBL alumnus Werner Kühlbrandt. Scientists from around the world have used cryo-EM to investigate the structure of biologically important molecules, as well as disease threats such as viruses.
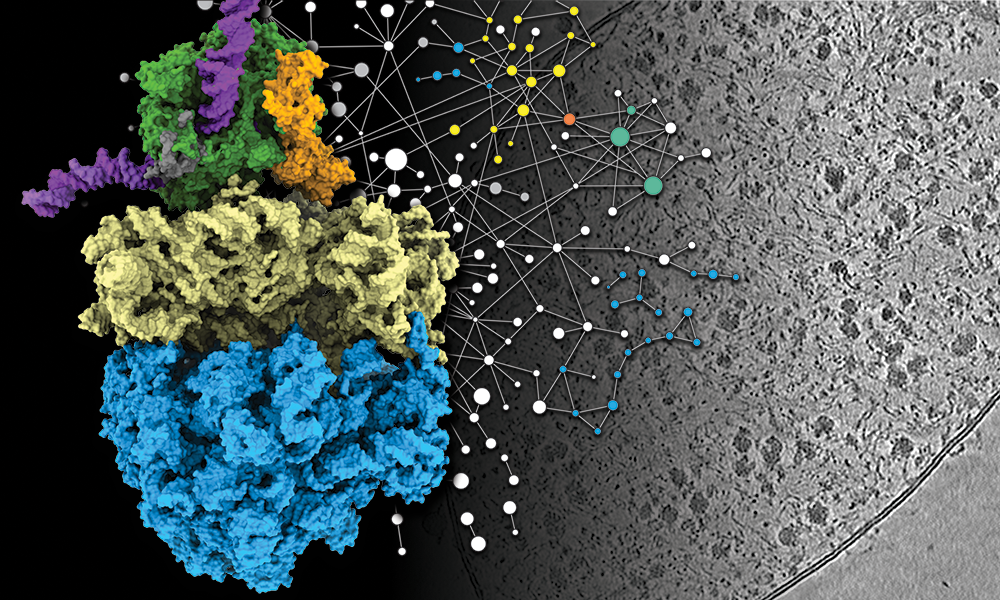
Another technique that has seen extensive use at EMBL is cryo-electron tomography (cryo-ET). While cryo-EM allows scientists to observe the structure of biological molecules, it usually still requires samples of carefully isolated molecules taken out of their cellular context. However, cryo-ET allows scientists to take snapshots of intact cells, along with all their internal components, which can later be reconstructed in 3D. In recent years, EMBL researchers have used cryo-ET to ask a number of important biological questions, including how antibiotics bind their targets, e.g. in ribosomes, and how this binding affects the functioning of the ribosomal machinery.
Databases and AI
What macromolecular crystallography, cryo-EM, and cryo-ET have in common is not just their potential for revolutionising our understanding of life at the atomic level, but also their propensity to generate huge volumes of data, which can be mined for insights. In the 1970s, with the rise in the number of protein structures decoded experimentally, it became essential to systematically store and share these data with other scientists. In 1971, the Protein Data Bank (PDB), was established as the central archive for experimentally determined macromolecular structure data. In 1999, EMBL-EBI researchers helped set up the PDBe (Protein Data Bank in Europe), working with the PDB in the USA. Their aims were to help process the increasing number of structure depositions, validate depositions, and develop an extensible database derived in part from the PDB and operating under the worldwide Protein Data Bank (wwPDB) international collaboration, established in 2003.
In 2002, EMBL-EBI founded the electron microscopy data bank (EMDB), a public repository for cryogenic-sample Electron Microscopy (cryo-EM) volumes and tomograms of macromolecular complexes and subcellular structures. This was accompanied by the Electron Microscopy Public Image Archive (EMPIAR), a public resource for raw images underpinning 3D cryo-EM maps and tomograms launched in 2014. Similarly, the SASBDB is a fully searchable, curated repository of freely accessible and downloadable data related to small-angle scattering experiments. Such databases have been essential for the advancement of electron microscopy technology and standards, as well as transparency and validation efforts.
Similarly, PDB depositions have seen a rapid increase since the beginning of the 1990s, due to the numerous technological innovations in all major structural biology techniques, including improvements in instrumentation, analytical software, robot-assisted automation, and availability of high-brilliance synchrotron radiation and X-ray free-electron laser sources, enabling today not only high-quality experiments on increasingly challenging systems but also time-resolved studies. Over the last 50 years, EMBL has also contributed many computational tools for structural biology to handle, validate, characterise and display protein structures and their ligands, which are used worldwide.
The 130,000 experimentally determined protein structures in the PDB played a critical role in the development of one of the most exciting breakthroughs in structural biology in recent years – the artificial intelligence (AI) based protein structure prediction tool AlphaFold2. AlphaFold tackles a long-standing problem in structural biology – given a protein’s sequence of amino acids, how accurately can we predict its three-dimensional structure once it folds?
In 2021, DeepMind, the company which developed AlphaFold partnered with EMBL-EBI to make the AlphaFold protein predictions, source code and methodology freely and, crucially, openly available to the global scientific community through the AlphaFold database. In a major update the following year, the database expanded to include more than 200 million protein structures, covering almost every organism on Earth that has had its genome sequenced.
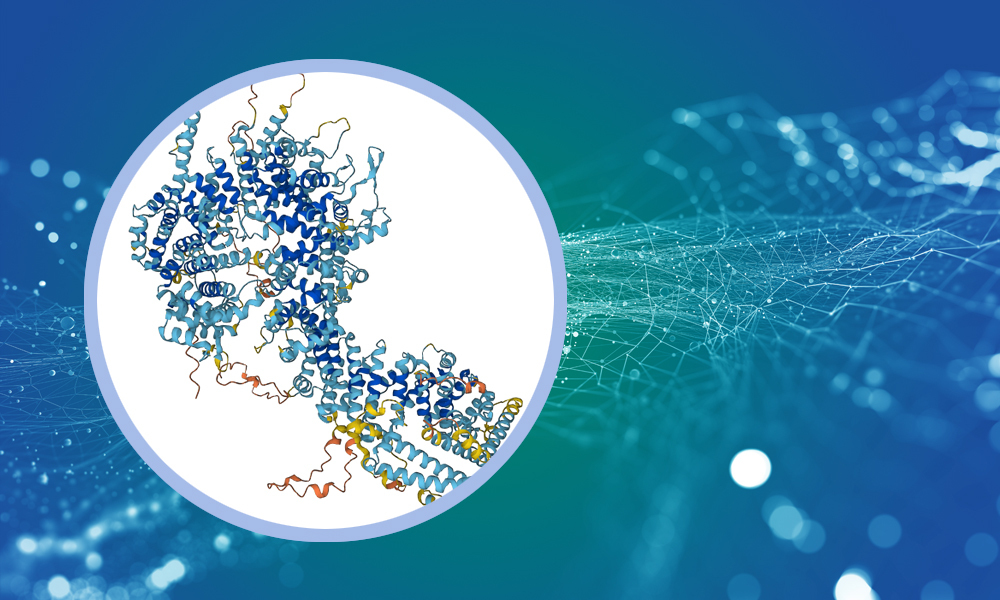
The availability of reliable three-dimensional models of proteins has sparked a revolution in the design and execution of experimental structural biology projects. It has also taken structural biology from a specialised field to something that can be part of almost every life science research project – it has prompted scientists who might not have otherwise ventured down structural biology lanes to ask “What does my protein of interest actually look like?” Further, projects like AlphaFold-Multimer and AlphaPullDown are being used successfully to figure out which proteins or peptides can interact with each other, with experimental validation as the subsequent step. AlphaFold predictions, therefore, provide a solid foundation for generating hypotheses about molecular function, which in turn propose mechanisms of action and enable the design of experiments with specific expected outcomes. This lets us investigate complex systems that would be difficult or impossible to study through conventional methods.
AI is also becoming increasingly important for cryo-EM/ET, helping with higher throughput and better-targeted data acquisition, reducing noise in data, annotating datasets, and more.
Looking towards the future
Using such an array of structural biology tools, EMBL researchers have shed light on some of the most fundamental mechanisms in living systems, as well as on those with strong relevance in human health. For example, in recent years, former and current EMBL researchers have provided key insights into the assembly of nuclear pores and a group that forms part of one of EMBL’s many different partnerships, the Molecular Medicine Partnership Unit in Heidelberg, has elucidated the way the human immunodeficiency virus (HIV) assembles. Other researchers have expanded our understanding of the mechanisms of transcription and translation, DNA remodelling and chromatin structure, drug and small-molecule transport across membranes, molecular interactions in pathogens, and more.
In addition to such seminal research, one of EMBL’s key roles in the last five decades has been in democratising access to structural biology techniques and data, so that structural biology is no longer a field confined to structural biologists. As we head into the next fifty years, we expect this trend to continue. Today, the boundary between structural biology such as crystallographic approaches, cryo-EM/ET, and what is classically considered imaging is fading away, and we are increasingly handling biology across scales, from millimetre-scale organisms and tissue through single cells to the nanometer scale of macromolecular complexes. In addition, we are increasingly seeing the incorporation of the fourth dimension – time – into structural biology experiments, allowing us to do time-resolved studies with crystallography and cryo-EM, with the potential to provide major breakthroughs following further technological developments. This is made possible with the increase in temporal resolution provided by techniques such as nuclear magnetic resonance (NMR), SAXS, and single-molecule FRET.
We also expect to see more in situ structural biology across scales and organisms, combined with mechanistic insight from biophysical and biochemical approaches, as well as further glimpses into the cell’s ‘dark matter’ – intrinsically disordered proteins, which do not have the traditional rigid 3D structures we usually associate with proteins.
Finally, we also foresee a systems structural biology approach in the future, where rather than one structure at a time, we will learn about ensembles of structures of the same molecular assembly, or even about multiple assemblies at once with cryo-ET of intact cells or in complex reconstitutions. This is where EMBL’s other strengths in bioinformatics and data integration will be a powerful combination with structural biology.
Learn more about EMBL’s contribution to life science research and services in our 50th anniversary commemorative publication.


