Fighting bacteria – with viruses
Scientists show how bacteriophages destroy Clostridium difficile cells, opening up new possibilities for using viruses as an alternative to antibiotics.
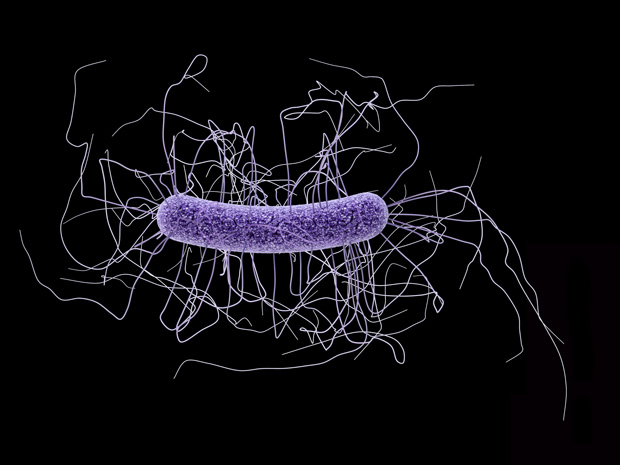
Research by scientists from EMBL Hamburg reveals how enzymes from bacteriophages – viruses that infect and destroy bacteria, but do not affect other organisms – are triggered and released to degrade cell walls of the bacteria Clostridium difficile (C. diff). Published today in PLoS Pathogens, the work adds crucial information to our understanding of the bacteriophage infection pathway, and opens up new opportunities for developing effective therapies to overcome issues raised by antibiotic resistance.
Side-stepping antibiotics
C. diff, is becoming a serious problem in hospitals and healthcare institutes, where it can cause life-threatening cases of diarrhoea. Patients who receive broad-spectrum antibiotic treatment are particularly at risk. C. diff naturally occurs in human gut flora and poses no problem in healthy individuals, but in patients treated with antibiotics, a large amount of gut bacteria are wiped out allowing the more resistant and persistent C. diff to increase uncontrollably in number, leading to complications. Such cases are very difficult to treat, precisely because C.diff is unresponsive to many antibiotics.
A potential alternative treatment for C. diff infections would be to use bacteriophages. These bacteria-infecting viruses were discovered as treatment for bacterial infections over 100 years ago but became less popular as antibiotics – which were easier to use and store – became available. Now, with the increase in antibiotic resistance, bacteriophage research and therapies are experiencing a revival.

Bacteriophages enter a bacterial cell and hijack that cell’s DNA replication machinery to reproduce. The cell then breaks up, releasing the newly formed bacteriophages. In order to develop and engineer effective bacteriophage therapies, a clear understanding of the viruses’ life cycle is needed – in particular, how the bacterial cell wall is destroyed. While it is known that the enzymes involved, called endolysins, are produced at the end of the bacteriophage life cycle directly before the break-up of the cell, just how these enzymes are activated remains a crucial missing part of the puzzle. Understanding this mechanism would allow researchers to engineer effective bacteriophage treatments.
A common switch
In this study, Rob Meijers and his group at EMBL Hamburg report a common activation mechanism for bacteriophage endolysins that target Clostridium bacteria like C.diff. In collaboration with Melinda Mayer and Arjan Narbad from the Institute of Food Research in Norwich, UK, the scientists compared two endolysins. One was retrieved from a bacteriophage that infects C. diff, and the other digests the cell wall of a Clostridium species that impairs cheese fermentation. At the German Electron Synchrotron (DESY) in Hamburg, the EMBL researchers used X-ray crystallography and small angle X-ray scattering – techniques which involve shining X-ray beams on a sample and measuring how that sample interferes with those rays – to deduce the enzymes’ 3-dimensional structure. From that 3D structure, Meijers and colleagues were able to infer how the endolysins work.

Credit: EMBL/ROB MEIJERS
“These enzymes seem to take on two different conformations,” explains Matthew Dunne, a PhD student in Meijers’ lab who carried out the research. “They appear to switch from a tense, elongated shape where a pair of endolysins are joined together, to a relaxed state where the two endolysins lie side-by-side.” The switch from one state to the other triggers the release of the active enzyme, which then begins to degrade the cell wall. Once the cell wall begins to break down, the bacterial cell can no longer withstand its own internal pressure and explodes, releasing the new bacteriophages that go on to infect other bacteria. The group believes environmental influences trigger the release and activation mechanism.
“Remarkably, we found that the two endolysins have a common activation mechanism,” explains Meijers. It therefore seems likely that this mechanism is not restricted to Clostridia-specific bacteriophages, but can be found in many others, too. “This knowledge could allow us to engineer effective, specific bacteriophages, not just for C. diff infections, but for a wide range of pathogenic bacteria related to human health, agriculture and the food industry. In the light of increased antibiotic resistance, bacteriophages and their endolysins may provide a good alternative.”