
Team spirit in the genome
Genes, like people, are fundamentally social. Just as we often work in teams, companies, or other more or less complex organisations, genes often work together in genetic networks. And just as our productivity is often influenced by who we work with, the effects of genes depend on the peers they interact with.
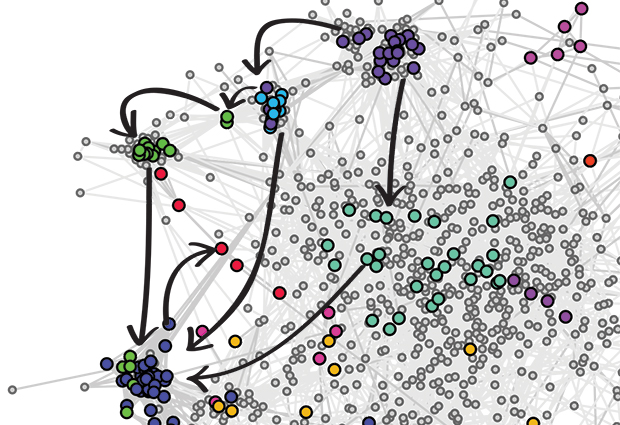
That’s why understanding genetic predispositions remains a challenge – each person’s genome is a unique combination of genes, and it’s difficult to work out how they will interact and function as a team. In football, a team of star players may end up standing in each other’s way, whereas a team with good team spirits can achieve success that one might not expect from the players individually.
For the past 100 years geneticists have tried to untangle the complex webs of genetic interactions, and to identify how gene variants affect what other genes do. Ideally, biologists would like a global picture of all genetic interactions in the cell, but this has been hard to track down. Now work led by Michael Boutros of the German Cancer Research Centre (DKFZ), Heidelberg, in collaboration with Wolfgang Huber at EMBL Heidelberg, has shown how it can be done. The study is reported today in the journal eLife.
The team developed their approach using cells from the fruit fly Drosophila. First they selected genes which, when mutated, had an effect on important characteristics like cell growth and division. This generated a list of 1367 genes, of which 72 were picked out as likely hubs in the genes’ social network.

To work out which of these genes – or, more precisely, the proteins they produce – interact, the team set about silencing pairs of genes using a technique called RNA interference (RNAi). The logic behind this approach is that if the effect of silencing both genes at the same time is different from what is expected from the effects of silencing each of them singularly, then that points to a genetic interaction. In these RNAi experiments, each of the 1367 genes was silenced in combination with one of the 72 key genes. “We took more than a million images of cells”, says co-author Thomas Sandmann of DKFZ, “and tested almost 100 000 pairwise combinations of silenced genes.” All this data was analysed with automated algorithms on a bank of computers, a process that Bernd Fischer, formerly a member of the Huber lab and now at DKFZ, says “would have taken more than two years on a single computer”.
We took more than a million images of cells and tested almost 100 000 pairwise combinations of silenced genes
Thirteen per cent (12,361) of the pairs showed evidence of a genetic interaction, indicating that they work together. But Fischer and his co-workers weren’t just content knowing which genes interact – they wanted to know how. So they developed a method to work out the direction of genetic interactions – whether gene A influenced gene B, or vice versa. “This is novel, and hasn’t been done on this scale before,” says Huber. Beyond revealing the direction of the interactions, their analysis also showed whether genes amplified, or diminished, the effects of each other. This way of teasing out the way genes interact across the whole cell could be used to shed light on the genetic interactions in many complex cellular processes, from fruit flies to human cells.
Such insights will eventually help towards gaining a better understanding of genomes and their output – what biologists call the phenotype – and also for finding new targets for anticancer drugs. Typically, it is difficult to restore the function of proteins that are broken by mutations in cancer, but maps of genetic interactions provide a way around that problem. “New drugs attempt to exploit lethal genetic interactions to specifically target vulnerabilities in cancer cells,” says Boutros. “And genetic interactions may also explain how resistance to cancer drugs arises.”


