
3D-Beacons Network: protein structure data, all in one place
3D-Beacons Network acts as a one-stop shop for protein structures by combining and standardising data from several providers
A new platform called 3D-Beacons Network brings together experimentally determined and predicted protein structure models and related data from several providers and makes them freely available, all in one place.
The data are provided by the Protein Data Bank in Europe (PDBe), AlphaFold DB, SWISS-MODEL, Protein Ensemble Database (PED), Small Angle Scattering Biological Data Bank (SASBDB), Genome3D, and PDBe Knowledge Base (PDBe-KB).
Proteins are the essential building blocks of all living organisms, and understanding their structure is critical for advancing medical research. Protein structures are traditionally determined using complex lab-based techniques. However, with the availability of high-quality predicted models for large numbers of proteins, structural biologists can now access both experimental and predicted structures together with their annotations, to benefit their research.
The 3D-Beacons Network allows users to search using UniProt accession numbers and see protein structure models from different providers in a standardised format. This collaboration combines experimentally determined structures, predicted models, template-based models, conformational ensembles, and annotations.
Standardised data
“Until now, users had to go to many different resources to find out whether there are protein structures available and then access those data,” said Sameer Velankar, PDBe Team Leader at EMBL-EBI. “With 3D-Beacons, they can query everything they need in one place, and the platform will standardise the way they access the data.”
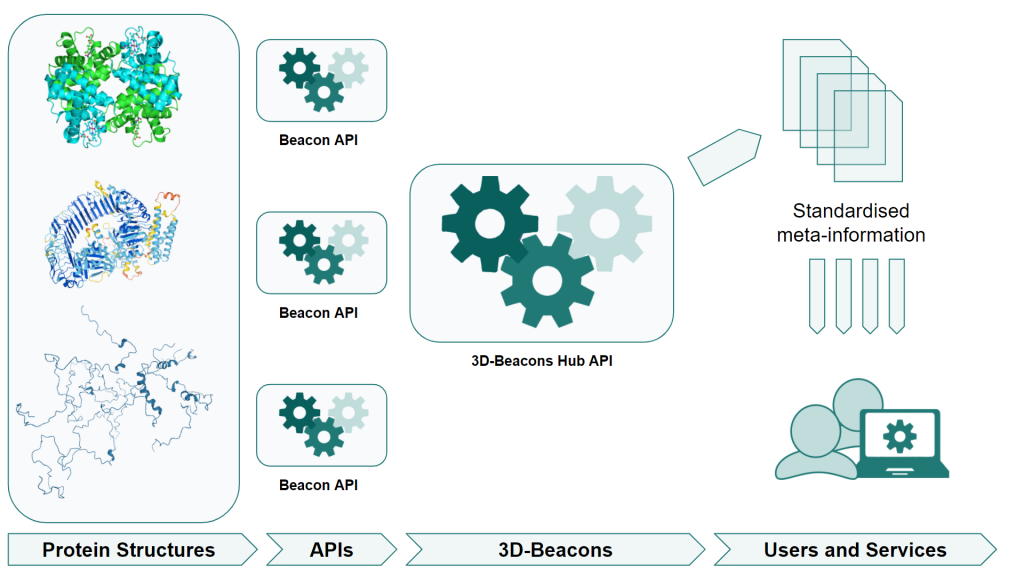
The 3D-Beacons Hub, hosted at EMBL-EBI, acts as the centre of the 3D-Beacons Network. Within the Hub, a set of application programming interface (API) endpoints provide options to query the different types of data from different providers. The 3D-Beacons Hub API collates and standardises these data so users of the 3D-Beacons Network can access them in a common format. The 3D-Beacons Hub API is freely available through GitHub.
“3D-Beacons is a step change for the more than 70,000 scientists, students, and schoolchildren who use the Jalview application to understand proteins, their function, and evolution. The API makes it easy for Jalview users to discover and explore the latest 3D structures and predicted models for proteins,” said Jim Procter, Coordinator of Jalview at the University of Dundee.
What are APIs?
An API – or application programming interface – is a software intermediary that allows two applications to talk to each other in a standardised way. APIs let users get data using predefined URLs, without needing technical information of the data providers.
Oligomeric and mutant protein structures
”Researchers may wonder what extra data 3D-Beacons will be able to provide,” said Mihaly Varadi, a project leader at EMBL-EBI. “By collecting data from all our different providers, it’s possible to obtain specific protein structures, including predicted structures of oligomeric and mutant proteins, through 3D-Beacons.”
Structures of oligomeric proteins – which contain more than one subunit – and mutant proteins are currently scattered across different data resources. 3D-Beacons will standardise access to these data.
“It will be incredibly valuable for researchers to have access to all this information in one place. For example, knowing the location of a mutated residue on a protein structure and whether there are known or predicted functional sites nearby can help in explaining whether that mutation is likely to be disease-causing,” said Christine Orengo, Professor of Bioinformatics at University College London.
Funding
Funding for this project was provided by the BBSRC.


