DNA’s twisted communication
The organisation of the genome is a key element for the control of gene expression
In a nutshell:
– Fgf8 is a crucial gene for embryo development; it is itself controlled by multiple and interdependent regulatory elements
– The pattern of expression of Fgf8 depends on the combination of the information conveyed by its various regulatory elements
– The communication between regulatory elements is mediated by their relative distribution along a chromosome rather than by their sequences
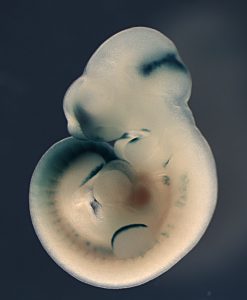
During embryo development, genes are dynamically, and very precisely, switched on and off to confer different properties to different cells and build a well-proportioned and healthy animal. Fgf8 is one of the key genes in this process, controlling in particular the growth of the limbs and the formation of the different regions of the brain. Researchers at EMBL have elucidated how Fgf8 in mammal embryos is, itself, controlled by a series of multiple, interdependent regulatory elements. Their findings, published today in Developmental Cell, shed new light on the importance of the genome structure for gene regulation.
Fgf8 is controlled by a large number of regulatory elements that are clustered in the same large region of the genome and are interspersed with other, unrelated genes. Both the sequences and the intricate genomic arrangement of these elements have remained very stable throughout evolution, thus proving their importance. By selectively changing the relative positioning of the regulatory elements, the researchers were able to modify their combined impact on Fgf8, and therefore drastically affect the embryo.
“We showed that the surprisingly complex organisation of this genomic region is a key aspect of the regulation of Fgf8,” explains François Spitz, who led the study at EMBL. “Fgf8 responds to the input of specific regulatory elements, and not to others, because it sits at a special place, not because it is a special gene. How the regulatory elements contribute to activate a gene is not determined by a specific recognition tag, but by where precisely the gene is in the genome.”
Scientists are still looking into the molecular details of this regulatory mechanism. It is likely that the way DNA folds in 3D could, under certain circumstances, bring different sets of regulatory elements in contact with each other and with Fgf8, to trigger or prevent gene expression. These findings highlight a level of complexity of gene regulation that is often overlooked. Regulatory elements are not engaged in a one-to-one relationship with the specific gene that has the appropriate DNA sequence. The local genomic organisation, and 3D folding of DNA, might actually be more important factors that both modulate the action of regulation elements, and put them in contact with their target gene.
More research will be necessary to understand in detail the impact of the 3D structure of DNA on the communication between the various elements of the genome, and on the regulation of gene expression. Further down the line, this could also further our understanding of how genomic rearrangements might disrupt these 3D regulatory networks and lead to diseases and malformations.