Enzymes’ superimposable structures revealed
EMBL scientists superimpose structures of two-metal-ion enzymes and reveal new potential drug targets
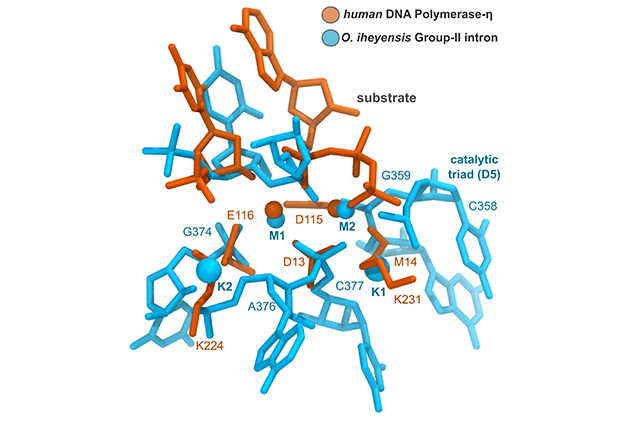
Marco Marcia, Group Leader at EMBL Grenoble, in collaboration with researchers from Istituto Italiano di Tecnologia (Italy), published a study today in Structure providing new understanding of so-called two-metal-ion enzymes. This diverse group of enzymes, which perform a great number of different tasks, all rely on two essential metal ions in their active site to synthesize and cut DNA and RNA, thus ensuring the correct flow of genetic information from genes to proteins. First author Matteo Colombo (Marcia lab) gives us more insight into the paper.
What is new about this study?
Our study shows that the similarities between two-metal-ion enzymes go beyond them having metal ions positioned at the same place in the active site. Although belonging to different enzymatic families, we knew that two-metal-ion enzymes follow analogous reaction chemistry, which reflects the similarity in their structures. In this study we also found two amino acids around the active site whose positions are also highly conserved. Such conserved amino acids could be involved in positioning the enzyme correctly to facilitate its work. We think this because when these positions are mutated the enzymes lose function. These observations improve our understanding of the reaction mechanism of these crucial metalloenzymes.
Why is the study so surprising?
Two-metal-ion enzymes belong to different enzymatic families that are functionally and evolutionarily different. Having evolved through separate paths, it’s fascinating that two-metal-ions adopt the same structure for their active sites and the region around them. In addition, our work was stimulated by a previous project in which we determined the structure of an RNA two-metal-ion enzyme, demonstrating the informative potential of RNA.
What could be a future application?
Our paper concerns enzymes that are also the focus of important biological and pharmacological research. Identifying new structural sites that are likely to be important for the enzyme’s activity could reveal promising new targets for drug development (for example, to inhibit nucleic acid synthesis in pathogens like influenza virus) or to expand the engineering of CRISPR/Cas9 for more specific, selective gene editing.


