How transcription factors work together
EMBL researchers have developed a method to observe interactions between transcription factors
Researchers in the Zaugg group have developed a statistical learning framework that allows them to study cooperativity between transcription factors, as they report in Nature Communications. The results of the study show that cooperativity between transcription factors from different families is mediated by specific DNA sites. This provides an additional regulatory step in DNA transcription.
A large set of possibilities
“It often is not just one transcription factor that regulates a gene. They usually act in cooperation,” says group leader Judith Zaugg. The function of transcription factors can vary strongly, depending on the context in which they act. “The same transcription factor, interacting with another partner, can be involved in a completely different type of disease. Changing the partner will have a functional impact,” Zaugg explains.
Ignacio Ibarra, postdoctoral fellow in the Zaugg group and the main developer of the new framework, used large-scale genomics data from an in vitro experiment to predict cooperative binding between pairs of transcription factors. Following up on a specific pair of transcription factor families, Forkhead and Ets, and using data from chronic lymphocytic leukaemia patients, he found that co-expression of these factors allowed patients to be efficiently divided into groups of high and low expression of both transcription factors. High expression levels were associated with later commencement of treatment.
The cooperativity predictions were also validated in vitro in collaboration with EMBL’s Hennig group. “In computational studies, the validation often turns out to be the missing part,” says Zaugg. “This study is a very nice example of what’s possible thanks to the EMBL environment.”
Although many transcription factors act in pairs, Ibarra explains that the functionality of this pairing has not yet been extensively studied. “The question now is: how many biological events were missed because the analysis only focused on one transcription factor and not on pairs?” he says. The framework Ibarra has created is a first step towards a larger catalogue of possible transcription factor cooperation functions that can be described with available data.
Dancing at the binding sites
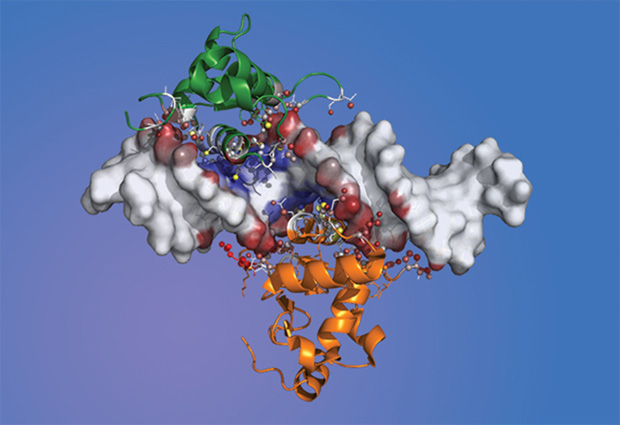
Ibarra observed that Forkhead–Ets pairs were not in direct contact with each other during their interactions, but connected at the DNA binding site through a protein residue. The dynamics between them reminded Ibarra of two dancers going back and forth, with DNA as the music that determines their movements.
“Transcription factor pairs are far less abundant than single transcription factors when you are observing data, but they are much more interesting,” says Ibarra. “You could picture a whole orchestra and single dancers, but in the middle of the floor there is a couple dancing that is drawing all the attention.”
Zaugg also likes this analogy, because it describes various aspects of the transcription factor interaction. “If you have a partner who dances well, you don’t need to dance so well yourself to look good,” says Zaugg. “And this is essentially what we found: one of the transcription factors binds very weakly and the other one very strongly, but still their interaction works. But without a partner, this dance is not possible at all.”
Wider benefits
It is planned to make the catalogue developed by the group available as a web service. The Zaugg group has several ongoing projects in which this tool will be valuable, and the huge amount of observable transcription factor interactions means that the scientific community at large will benefit from it. “It will be an important resource that researchers can use to have a look at their specific system and the specific transcription factors they are working with,” says Zaugg.
The Zaugg group also recently published diffTF, a piece of software that can be used by scientists to classify transcription factors into activators or repressors. “We are a computational group,” says Zaugg. “We work on projects that we find biologically interesting, but with a strong focus on computational tools, algorithms, or data generation that will prove useful to other researchers.”


