Getting wise to the influenza virus’ tricks
A high-resolution image of an influenza virus protein opens the way to design new antiviral drugs
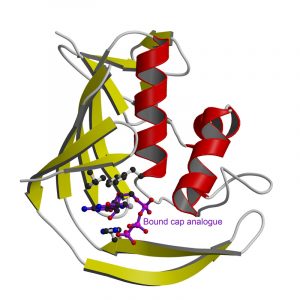
Influenza is currently a grave concern for governments and health organisations around the world. Now one of the tactics used by influenza virus to take over the machinery of infected cells has been laid bare by structural biologists at the EMBL, the joint Unit of Virus Host-Cell Interaction of EMBL, the University Joseph Fourier and National Centre for Scientific Research (CNRS), in Grenoble, France and at CSIC, Spain. In the current issue of Nature Structural and Molecular Biology they publish a high-resolution image of a key protein domain whose function is to allow the virus to multiply by hijacking the host cell protein production machinery. The findings open the way for the design of new drugs to combat future influenza pandemics.
Upon infection the influenza virus starts multiplying in the cells of its host. One protein that is crucial in this process is the viral polymerase – the enzyme that copies its genetic material and helps to produce more viruses. One component of the polymerase, called PB2, plays a key role in stealing an important tag from host cell RNA molecules to direct the protein production machinery towards the synthesis of viral proteins. Researchers of the groups of Stephen Cusack and Darren Hart at EMBL Grenoble have identified the PB2 domain responsible for binding the tag, produced crystals of it and examined them with the powerful X-ray beams of the European Synchrotron Radiation Facility (ESRF).
“Viruses are masters of cunning when it comes to hijacking the normal functioning of the host cell. The influenza virus steals a password from host messenger RNAs, molecules that carry the instructions for protein production, and uses it to gain access to the cell’s protein-making machinery for its own purposes,” says Cusack.
The password is a short extra piece of RNA, a modified RNA base called a ‘cap’, which must be present at the beginning of all messenger RNAs (mRNAs) to direct the cell’s protein-synthesis machinery to the starting point. The viral polymerase binds to host cell mRNA via its cap, cuts the cap off and adds it to the beginning of its own mRNA – a process known as ‘cap snatching’. The capped viral mRNA can then be recognised by the host cell machinery allowing viral proteins to be made, at the expense of host cell proteins.
The atomic resolution image the EMBL scientists generated of a PB2 domain bound to a cap reveals for the first time the individual amino acids responsible for recognising this special structure. The central interaction is a sandwich with two PB2 amino acids stacking either side of the cap. Whilst this recognition mechanism is similar to other cap-binding proteins, its structural details are distinct. Collaborators at the Centro Nacional de Biotecnologia in Madrid showed that disruption of the PB2 cap-binding site prevents the influenza virus from replicating.
“These findings suggest that the PB2 cap-binding site is a very promising target for anti-influenza drugs,” Hart says. “Our new structural insights will help us design mimics of the cap that would inhibit viral replication and hence reduce the spread of virus and the severity of the infection.”



