Nuclear pores in their natural context
The architecture of the nuclear pore complex differs inside cells compared to its form observed in vitro studies, and increases our understanding of crucial processes of life.
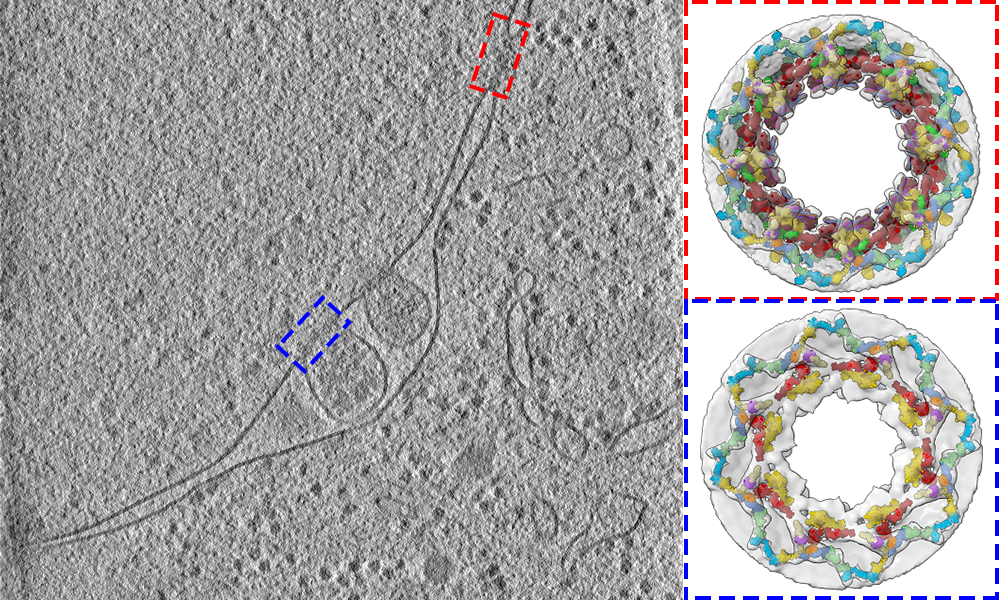
EMBL scientists study the 3D structure of nuclear pores in yeast cells
Scientists from the Beck group at EMBL Heidelberg, in collaboration with the Kosinski group at EMBL Hamburg and the Pfander group at the Max Planck Institute of Biochemistry in Martinsried, have studied the 3D structure of nuclear pores in budding yeast (Saccharomyces cerevisiae). Their findings, published in Nature, show how the architecture of the nuclear pore complex differs inside cells compared to its form observed in vitro studies, and increases our understanding of crucial processes of life.
Nuclear pores are a highly complex assembly of proteins. Thousands of them are embedded in the double membrane that surrounds and protects the cell’s nucleus. They act as a gateway that regulates the entry and exit of hundreds of thousands of molecules every minute. An important function of nuclear pores is to regulate the export of a molecule called messenger RNA (mRNA) from the nucleus into the surrounding cell – the cytoplasm – where it delivers instructions for the assembly of proteins.
Revealing the architecture
“We now appreciate better how the nuclear pore works in its native context, and what happens to the pore during physiological, pathological, or starved conditions,” says group leader Martin Beck, who led the structural work. The study provided a detailed structural description of the three protein rings that make up the nuclear pore, known as the cytoplasmic, nuclear, and inner rings. To show how these rings are arranged in cells, EMBL researchers used a combination of cell biology, computational modelling, and in-cell cryo-electron tomography: an imaging technique, under active development at EMBL, that is used to produce high-resolution 3D views of the molecular landscape inside a cell.
One of the study’s goals was to find out if the architecture of the nuclear pore would differ inside the crowded environment of a cell, compared to in vitro studies. Using cryo-electron tomography, the Beck group captured images of nuclear pores inside yeast cells. As they’d expected, the scientists discovered that some parts of the nuclear pore complex, when observed in cells, had different spatial configurations in comparison to previous in vitro studies. This led to fundamental new insights. “We found out that the 3D configuration of the cytoplasmic ring accommodates the path of mRNA export,” says Matteo Allegretti, a postdoc in the Beck group and first author of the study.
Understanding the life cycle
The structure of the cytoplasmic ring also serves another function – it exposes a certain domain of a protein to the cytoplasm. This domain interacts with proteins that facilitate the process by which nuclear pores are broken down by the cell and replaced with new ones – a process known as autophagic turnover. How exactly the architecture of nuclear pores facilitates the breakdown process – autophagy – and assembly of nuclear pores is largely unknown, but this study provides important first steps towards a better understanding of these mechanisms. “With knowledge coming from many different structures, we’re closer to understanding how nuclear pores assemble and how the pore evolved from the first cells with a nucleus up to now,” says group leader Jan Kosinski, who led the computational modelling.
To better understand the assembly of nuclear pores, the researchers grew a yeast strain missing a protein called nucleoporin 116, which plays an important role in the assembly process. The resulting structure was missing the cytoplasmic ring and part of the inner ring. The scientists conclude that these incomplete structures show intermediate states of nuclear pore assembly. Studying this process is important, as failures in the assembly of nuclear pores lead to the death of the cell and have been linked to neurodegenerative diseases. The study yielded detailed structures that scientists from other institutions can use in various ways, for example to study nuclear pore function, how molecules are transported into or out of the nucleus, or how viruses enter the nucleus. Many viruses, such as influenza and SARS‑CoV‑2, need to get their genetic information past the nuclear pore complex to infect a cell. “It also shows the scientific community that we need to shift scientific efforts towards the investigation of the structure–function relationship of macromolecules directly inside the cell,” says Matteo. Fundamental processes of life, such as nuclear transport and autophagy, can be understood by combining technologies like cryo-electron tomography with structural modelling, light microscopy, and biochemistry.



