AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science 10 June 2022
10.1126/science.abm9506
Combining AlphaFold2 with experimental and computational techniques has helped scientists figure out the human nuclear pore complex’s architecture in greater detail than ever before
The human nuclear pore complex (NPC) is a true molecular giant, which sits on the membrane separating the nucleus from the cytoplasm. It’s doughnut-shaped and works as both a gateway and a checkpoint for molecules that travel between the cytoplasm and the nucleus. Thereby, the NPC facilitates fundamental processes in the cell, such as gene expression and translation. The nuclear transport system also plays a role in several diseases, including neurodegenerative disorders, cancer and viral infections.
What’s the NPC’s structure? How are its proteins glued together? How does it attach to the nuclear membrane? These and other questions have now been answered by the Kosinski Group at EMBL Hamburg and Centre for Structural Systems Biology (CSSB), the Beck and Hummer Labs at the Max Planck Institute of Biophysics, and collaborators. They created the most complete model of the human NPC to date by combining the protein structure prediction program AlphaFold2 with techniques such as cryo-electron tomography, single particle cryo-EM and integrative modelling.
For structural biologists, the human NPC is a challenging yet exciting 3D puzzle, with around 30 different proteins each present in multiple copies. This amounts to around 1000 puzzle pieces, which form a round core with surrounding flexible parts. Until now, the most accurate models of the human NPC core only covered 46% of the structure. But now, building on two decades of previous research in the field, scientists have created a new model of the NPC structure that covers more than 90% of its core.
While previously proposed NPC models had gaps and contained some proteins only in fragments, the new model removes much of this ambiguity.
“It’s like when you disassemble and reassemble an electronic device. There will be always some screws left, and you just don’t know where they are supposed to be,” said EMBL Group Leader Jan Kosinski, who co-lead the investigation. “We finally managed to fit most of them, and now, we know exactly where they are, what they do, and how.”
How did the scientists achieve this? The key was to combine several experimental and computational methods. This enabled the scientists to visualise the NPC at different scales and levels of detail.
For example, to model the overall silhouette of the NPC, the researchers used cryo-electron tomography. With this technique, they were able to observe the NPC in its cellular environment, rather than in isolation. More details of the individual protein building blocks were revealed by AlphaFold2, an artificial intelligence-based program that predicts protein structures, created by the company DeepMind.
“AlphaFold2 was a breakthrough moment for us,” said Agnieszka Obarska-Kosińska, postdoc who performed the molecular modelling. “Before, we didn’t know the structure of many proteins within the NPC. You cannot assemble a puzzle when you don’t know what the pieces look like. But AlphaFold2 combined with other approaches enabled us to predict those shapes.”
To refine the picture even further, the researchers used ColabFold, a version of AlphaFold2 modified by the scientific community to model interactions between proteins. This allowed them to visualise how the different puzzle pieces combine to form smaller subcomplexes, and how those subcomplexes are then glued together to form the NPC.
Finally, they put all the pieces together using the software Assembline previously developed by the Kosinski Group, and validated it against experimental data.
The resulting model was so complete and detailed that it enabled the researchers to create time-resolved molecular simulations that explain how the NPC proteins and the nuclear membrane interact to create a stable pore and how it responds to mechanical cues.
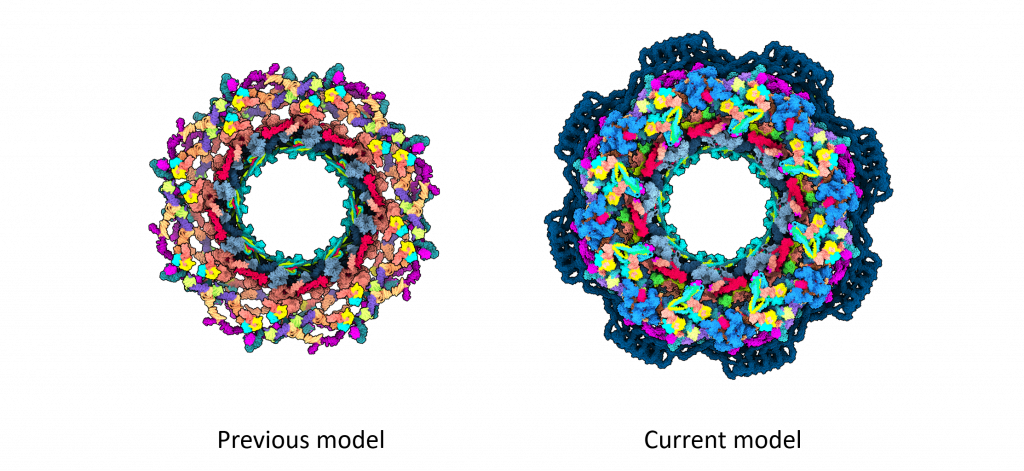
This work was a big leap forward for NPC research, but there is still a lot left to explore.
“This work exemplifies how, in the future, structural biology will embrace cell biology to create atomic models of ever larger assemblies of molecules that perform different functions in different parts of the cell,” said Martin Beck. Gerhard Hummer agrees: “We can now think of building a complete dynamic model of the NPC and simulate nuclear transport in atomic detail.”
The Kosinski Group will aim their future work at developing automatic methods for integrating structural and microscopy data using AlphaFold2 and their own software Assembline. They plan to apply these approaches to studying molecular processes driving viral infections.
Studying the NPC in its cellular surroundings also aligns with the goals of the new EMBL Programme ‘Molecules to Ecosystems’, which aims to explore life in context.
Science 10 June 2022
10.1126/science.abm9506