
Seeing deeper inside cells
EMBL scientists are probing the smallest parts of cells for answers to the biggest challenges

The lab looks like many others. PhD students and postdocs alternate between staring deeply at computer screens and gliding office chairs across the floor to consult with a colleague. There’s the concentrated hush of scientists at work. But the quiet belies the energy and excitement that members of the Mahamid group convey as they talk about their work and the technology they’re helping to shape.
“Complex biology can be explained by multifaceted but simple interactions, and what this technology provides seems like magic,” says Xiaojie Zhang, a postdoc who uses the technology the group is quickly developing and mastering to study stress granules – a type of structure involved in the cell’s stress response – in HeLa cells. She laughs. “The reason I get excited is that seeing is believing, and we can potentially see at near atomic resolution how molecules interact, directly in their biological context.”
While cryo-electron tomography (cryo-ET) was first envisioned in 1968, the advances the Mahamid group are bringing to this 3D method for studying molecules directly inside cells are new, and are likely to greatly expand its use. A tour through their projects hints at solutions to molecular puzzles and promises a future with innovative automated technology that will change the way biologists think about cells.
“These past few years have been transformative for those of us engaged in cryo-ET,” says group leader Julia Mahamid. “We’re at the point of pushing what was once thought to be a more boutique method to become a high-throughput one. Two years ago, we didn’t think this kind of capability was possible. Now, here we are.”
Julia first began working with cryo-ET as a postdoc at the Max Planck Institute of Biochemistry in Martinsried, near Munich. Her mentor was Wolfgang Baumeister, who pioneered key aspects of cellular cryo-ET. Since 2017, Julia has led her own group at EMBL, working with them to further develop and refine this technique.
Cryo-ET evolving
Cryo-ET piggybacks onto another state-of-the-art technique, cryo-electron microscopy (cryo-EM), which emerged from the pioneering work of Nobel laureate Jacques Dubochet at EMBL in the 1980s. Cryo-EM involves rapidly freezing samples at very low temperature and imaging them using a beam of electrons. By freezing samples, it’s possible to reduce the damage caused by the electron beam, and the rapid freezing process prevents the formation of ice crystals, which would damage and distort the fine structure of molecules.
Scientists can use cryo-EM to study biological structures at the atomic level. It’s possible to glimpse structures in high resolution, but this requires scientists to create highly purified samples taken out of their natural context of the cell. The age of cryo-ET seized on technical advances in electron microscopes, detectors, and computational analysis to create something akin to a CT scan for cells. It uses some of the same instruments as cryo-EM, but expands what scientists can see and do.
With cryo-ET, scientists can produce 3D snapshots of a cell to observe the activity and interactions of the molecules inside it at the highest resolution. Researchers prepare specimens by growing cells on a thin gold or titanium grid overlaid with a thin film that supports electron microscopy samples. They rapidly freeze the grid and then use a focused ion beam to shave away surplus material bit by bit, producing a thin layer, or lamella, around 200 nanometres (one five-thousandth of a millimetre) thick. A critical factor is that the cells are unperturbed, frozen in time, and the molecules within them are visualised in situ – in their natural habitat – having experienced a minimum of interference.
“At the moment, the skill set required to do cellular cryo-ET is fairly unique, but efforts towards automation will streamline the processes and make it a far more routine, push-button technique,” Julia says. “Scientists will be able to spend far more time thinking about experimental design than how to get their samples imaged.”
Mauricio Toro-Nahuelpan, a postdoc in the Mahamid group, recalls his work of only three or four years ago when automation was still just a dream and he’d need to be in the lab for 72 hours straight, babysitting his samples so he could manually reposition them to capture the high-resolution imagery.
While samples ultimately need to be about 200 nanometres thick, they might begin around 5,000 nanometres (one two-hundredth of a millimetre) in the case of yeast cells, or even bigger in other samples. Scientists can shave away or ‘mill’ the sample to achieve the thin lamella. Its thinness allows them to capture high-resolution images inside the cell.
Sara Goetz, a third-year PhD student in the group who uses cryo-ET to study how stressors impact yeast cellular structures and behaviour, helped to automate this process, along with Herman Fung – a shared postdoc with EMBL’s Müller group – and Sven Klumpe, a PhD student from the Max Planck Institute of Biochemistry. They created software with a graphical interface that enables scientists to pre-program the thinning process that they’d previously had to do manually. In addition, they worked with Wim Hagen, a senior engineer for EMBL’s Cryo-EM Service Platform, to make the imaging highly automated. Now, instead of taking only 12–14 tomograms over a weekend, with the automated system they can get at least 24 tomograms overnight. Additionally, group members can periodically check on the process remotely, and the microscope will email them if it encounters a problem. The scientists have also worked with Irene de Teresa – a shared postdoc with EMBL’s Zaugg group – to adapt deep learning techniques, enabling them to further automate data mining to locate molecules in tomograms and therefore in their native environments.
“There are so many technological limitations to this work, but this is no longer one of them,” Sara says. “This automation and high throughput mean we can tackle even more complex biological questions.”
When cells are placed on the grid used for creating cryo-ET images, they grow randomly across it. For the milling step, however, cells must be positioned at specific locations to be accessible for milling. To help with this issue, Mauricio has led the way in developing a micropatterned grid so cells can selectively grow at specific locations that have a suitable biosubstrate. Micropatterning not only provides a good substrate for culturing cells, but also ensures a surplus of cells available for the milling step, increasing the throughput. Micropatterning, together with the automation of the milling step by Sara and colleagues, is significantly helping to streamline the cryo-EM pipeline for cells.
“Micropatterning also allows us to create complex shapes, such as stars and crosses, on the grid where cells can grow,” Mauricio says. “In this way, cells adopt the shape of these patterns, rearranging their internal architecture. We can use these patterns to study cellular mechanical behaviour, expanding the scope of questions that can be addressed by cryo-ET.”
Layering techniques for better outcomes
Another group member, Edoardo D’Imprima, has tweaked the cryo-ET process to better pinpoint how, where, and when tumours first arise within a healthy tissue. His work involves organoids: 3D tissue cultures often described as mini organs because, even though they may not look like the organ they represent – mammary glands in the case of Edo’s work – they’re capable of the same functions.
Organoids are much larger and more complex than yeast cells, HeLa cells, or any other samples studied in the Mahamid group. To have a comprehensive structural understanding of organoids, one single imaging technique is not sufficient. Edo’s aim is therefore to develop an integrated pipeline that encompasses four other imaging techniques across different spatial scales – from millimetre-sized to near-atomic details.
Maintaining a frozen state without image-distorting formation of ice crystals is a major challenge for Edo in using cryo-ET for his research. To solve this problem, he applies high-pressure freezing, in which a specimen is exposed to liquid nitrogen but at much higher pressure (2,000 bar) to slow down ice formation.
“We have to ‘trick’ physics and use the high-pressure freezing approach,” Edo says. “That’s the beauty of water – it always gives you a chance to trick physics.”
Like Edo, other group members incorporate various observational tools to expand what they can see and learn from cryo-ET. Xiaojie, for example, combines live-cell imaging, cryo-confocal light microscopy, and cryo-ET in her research involving HeLa cells. To observe dynamic cellular structures, she uses genetically modified cell lines that produce fluorescent labels on the key component of the stress granules she is studying. By overlaying images produced by other methods, showing cells under stress at various time points, Xiaojie can more precisely navigate her cryo-ET images. Eventually, high-resolution structural information will help to provide a full perspective on how these dynamic structures assemble and disassemble, when cells respond to and are released from stress.
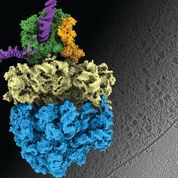
Earlier this year, Liang Xue, a PhD student in the group, and collaborators published results from a study involving Mycoplasma pneumoniae – a bacterium that causes a mild form of pneumonia. The research team combined cryo-electron tomography with cross-linking mass spectrometry and computer modelling to refine their findings, producing the highest-resolution images of cells ever obtained. The scientists were able to confirm that, in bacterial cells, which have no nucleus, the processes of transcription and translation – which cells use to convert information from DNA to proteins – can be coupled together, with the molecular machinery for these processes in physical contact. This had been a hypothesis for decades, but had never been confirmed. They also learned that the observed molecular activity in its natural context inside cells is different from what had been observed in previous studies outside the cell, further validating the importance of the in situ perspective.
“Image processing is a significant challenge. More than large datasets and heavy computation, we need new concepts and new algorithms,” Liang says. “It’s never easy and perfect, but that’s why we do it. With the new methodologies, we hope to lead structural biology into a new era: cellular, physiological, integrative, and systematic.”
Tomorrow’s cryo-ET
The innovative spirit isn’t lost on others outside the group, who look at the methods being refined and see how cryo-ET could expand their own research. In an interview earlier this year about his research on influenza, head of EMBL Grenoble Stephen Cusack explained how the work of the Mahamid group could help him to look directly at one of the flu virus’s key enzymes – influenza polymerase – and see it at work inside the nucleus of its host cell.
Many other scientists are excited by the potential of cryo-ET. Universities around the world, Julia explains, are investing in the technology so they too can make new leaps forward in understanding cellular structure and function. Cryo-ET has emerged as the most powerful technique for the structural understanding of biological molecules in their natural context.
“The big advantage of cryo-ET is how it opens up possibilities to discover new things,” Julia says. “You aren’t labelling one specific thing and only looking at what that thing is doing. Instead, you’re looking at everything at once, over a very wide range of spatial resolutions. The discovery potential is enormous.”
She acknowledges that this can be both a blessing and a curse, as it adds dimensions that bring a new complexity to the research. A scientist may begin with the intention of investigating one area but then encounter unanticipated new questions that challenge their assumptions. “Our current bottleneck is mining our data to get information from the images. Right now, we have 3D images, but we need to somehow convert them into a scientific understanding. This is where the field still needs to develop,” she acknowledges.
In the next 10 years, microscopes and detectors will continue to improve, but the computational side is where Julia sees the greatest likelihood for growth. In an era when artificial intelligence is rapidly developing, finding a way to incorporate AI, deep learning, and computer vision into image processing will be important. “There’s a state of mind that develops in working with cryo-ET,” Julia says. “Scientists working in this field – like those in my group – are excited about science and don’t see limitations. That’s true for all method developers: we can’t be restricted by what’s available. We are problem solvers. If a tool that we need doesn’t exist, we have to create it.”


