Structural basis for sequence-independent substrate selection by eukaryotic wobble base tRNA deaminase ADAT2/3
Nature Communications 8 November 2022
https://doi.org/10.1038/s41467-022-34441-z
EMBL Grenoble scientists provide new insights into the function of an essential RNA editing enzyme
A single cell in our body can contain all the genetic information for the whole organism. However, different types of cells require different sets of active genes to fulfil their specific functions. Gene expression – the process via which a cell selects necessary genes and uses this information to produce proteins – is thus critical to an organism’s health and normal functioning.
The Kowalinski group at EMBL Grenoble has recently solved the first structure of the active form of an important molecular player involved in gene expression. The molecule – an enzyme called ADAT2/3 deaminase – is found in almost every living organism. In a new publication in Nature Communications, the scientists demonstrate how this enzyme is able to recognise and edit a specific type of RNA to ensure the correct decoding of genetic information.
Gene expression is a highly regulated process, involving many crucial steps and active players. Any problem occurring along the way can lead to cellular deterioration and dysfunction, sometimes leading to disease. It involves two major steps. First, the cell copies the genes it actively needs from DNA to a molecule called messenger RNA (mRNA). Then, other molecular machines use the information in the mRNA copies to create a chain of amino acids arranged in a particular order to finally produce a protein.
Information encoded in the mRNA is stored in the form of three-letter words (‘AAA’, or ‘AAC’, etc.), called ‘codons’. Each letter represents a nucleotide – a chemical constituent of RNA – that can come in one of four varieties – A, U, C, and G. Each codon, in turn, codes for a particular amino acid.
To translate the information in mRNA into the language of amino acids, the cell uses a different type of RNA – transfer RNA (tRNA). tRNA serves as the physical link between the mRNA message and the protein output. All tRNAs have a characteristic ‘L’-shape, carrying a specific amino acid on one end and a codon-recognising sequence called the ‘anticodon’ on the other.
However, all tRNAs need to undergo extensive processing by cellular enzymes before they can perform this function. These enzymes can add small additional functional chemical groups at distinct positions, which are important for the correct folding of the L-shape and for flawless tRNA-mRNA matching.
The Kowalinski group is interested in how such chemical modifications can influence or change RNA function. “With our research, we want to answer specific questions: How exactly do enzymes ‘write’ an RNA modification or interact with it? How are these processes regulated? What are the consequences of RNA modifications?,” said Group Leader Eva Kowalinski. “Maybe, in the future, these enzymes could also be used as molecular tools.”
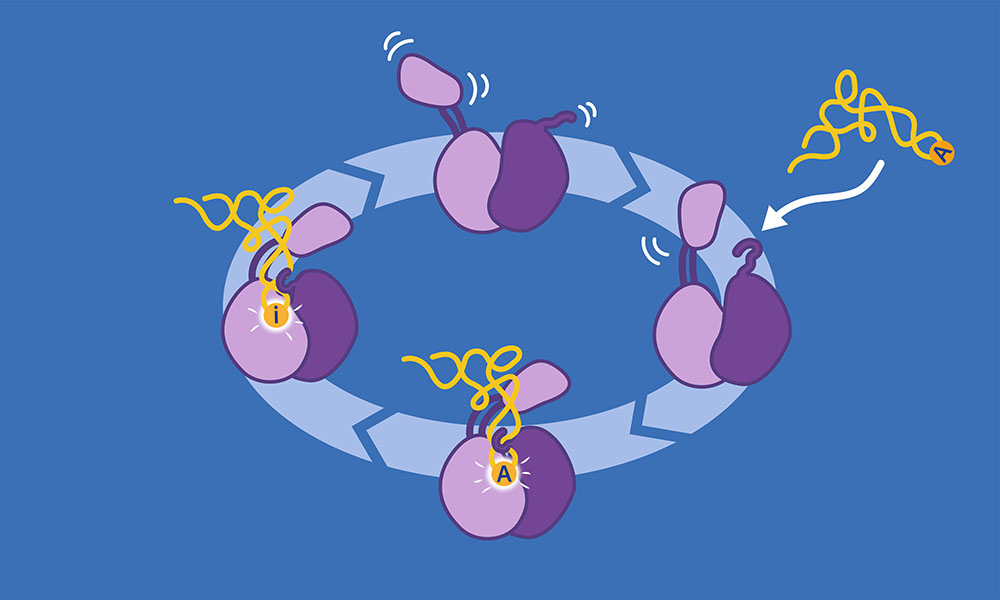
They are particularly interested in the modification of the tRNA’s anticodon sequence when the first letter is A (adenosine). In this case, it needs to be changed into another letter, inosine (I), which allows the tRNA to recognise a wider range of mRNA codons. This process is called ‘deamination’ and needs the intervention of a specialised enzyme called the ADAT2/3 deaminase.
ADAT2/3 deaminase has an important problem to solve — how to specifically recognise tRNAs among all the various other kinds of RNA molecules present in a cell. The Kowalinski group succeeded in obtaining the cryo-EM structure of ADAT2/3 and showing in detail how the different domains of the enzyme interact with a tRNA molecule to recognise its shape. In the process, they also obtained insights into the factors that may have driven the evolution of the enzyme.
In eukaryotes – organisms like animals and plants whose cells have nuclei – ADAT2/3 deaminase can edit up to 8 types of tRNAs. However, the enzyme can edit only one type of tRNA in bacteria and does not appear to exist at all in archaea – another supergroup of single-celled organisms.
“It is very interesting to analyse the structure of the enzyme from the evolutionary point of view,” said Luciano Dolce, postdoctoral fellow in the Kowalinski group and first author of the publication. “We have identified in the structure how two new domains of the enzyme interact with RNA. These only exist in eukaryotes and act like wobbling small arms able to recognise, catch, and guide the tRNA into the correct place. This shows how ADAT2/3 deaminase evolved to be a multi-tRNA receptor and editor, capable of recognising all tRNAs in eukaryotes and not go off target on other RNA species.”
The team collected data at EMBL Heidelberg, with help from experts in the Electron Microscopy Core Facility. Determining the enzyme’s 3D structure proved to be particularly challenging because of its small size and its ‘wobbly’ nature – the latter making it impossible to decode with macromolecular crystallography, another structural biology technique.
The team used ADAT2/3 deaminase extracted from the Trypanosoma brucei (T. brucei) parasite – an organism at the heart of the group’s research studies – to obtain the 3D structure. However, this study also opens up possibilities to look into the human equivalent of this enzyme, which the Kowalinski group predicts to be structurally similar with the one from T. brucei.
Since tRNA editing is so important to the cell’s functioning that tRNA pools co-evolved with ADAT enzymes, the team also plans to collaborate with the Boskovic group based at EMBL Rome, bridging structural insights with the roles of tRNA modifications in development and mammalian cell physiology.
Nature Communications 8 November 2022
https://doi.org/10.1038/s41467-022-34441-z