A quantitative map of nuclear pore assembly reveals two distinct mechanisms
Nature 4 January 2023
10.1038/s41586-022-05528-w
EMBL Heidelberg researchers and their collaborators reveal how one of the biggest molecular machines in eukaryotic cells is assembled one protein at a time
Fungi, plants, and animals have more in common than first meets the eye. Unlike archaea and bacteria, these organisms are ‘eukaryotes’, meaning that they store their genome in a dedicated cellular compartment called the nucleus. In a recent study, researchers from EMBL Heidelberg, the Max Perutz Labs in Vienna, and the University of California San Francisco have provided novel insights into how one of the key molecular machines in the nucleus is assembled.
The compartmentalisation of the genome in eukaryotes both safeguards the DNA and permits more intricate regulatory control over gene expression. But it also causes a logistical problem: to regulate gene expression in the nucleus and synthesise proteins outside of it, molecules must be able to cross the membrane that separates the nucleus from the rest of the cell.
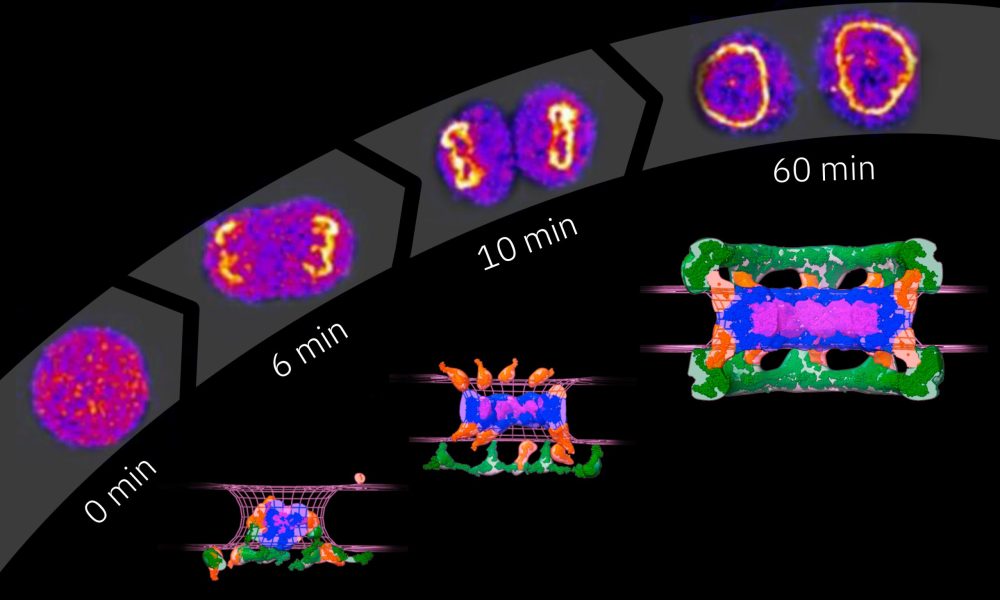
Nuclear pore complexes (NPCs) make such traffic possible, acting as gateways between the nucleus and the cytoplasm. On the molecular scale, these channels are giant-sized. Each pore consists of 8-48 copies of over 30 different proteins. Hundreds of individual proteins are thus required to build just one NPC, and thousands of NPCs are embedded in the nuclear envelope of a single cell. The new study from EMBL researchers and their collaborators provides the first molecular step-by-step guide to how all these components assemble into a functional pore.
“Due to the complexity of nuclear pore complexes and the fleeting nature of assembly events, it has been technically very challenging to study the assembly of these structures,” explained Shotaro Otsuka, the paper’s first author who pursued the project as a postdoctoral fellow in EMBL’s Ellenberg group before becoming a group leader at Max Perutz Labs. “Consequently, scientists have struggled to understand how these complex molecular machines are built.”
In higher eukaryotes, NPCs assemble around the nuclear envelope when it reforms after cell division, as well as when it expands in preparation for the cell to divide. To overcome the challenges associated with studying NPC assembly, the scientists utilised a quantitative fluorescence imaging approach developed in the Ellenberg group. The technique allows researchers to count over time the exact number of proteins involved in a specific process inside a living cell.
First, the scientists fluorescently tagged nucleoporins, the basic building blocks of NPCs. Using light microscopy, they then quantified how their number changes during the assembly of NPCs. The team had previously obtained structural models NPC assembly through electron microscopy, which they then integrated with this new data with the help of Andrej Sali’s lab at the University of California San Francisco. Using computational modelling tools and quantitative information on protein copy numbers over time, the scientists were able to propose a detailed structural model of NPC assembly post cell division.
“We were able to delineate the assembly order of the NPC in detail. Much to our surprise, we found that cells use two molecularly distinct assembly mechanisms: a rapid one that occurs directly after cell division, and a slower one that takes place when nuclei grow at a later stage of the cell cycle,” said Jan Ellenberg, EMBL group leader.
The data also revealed key mechanistic differences between the two assembly pathways. During post-cell division NPC assembly, the inner core of the pore – called the central ring – is built first. However, when NPC assembly takes place later in the cell cycle, the central ring is added only after other proteins – called nuclear filament proteins – are assembled.
Using this approach, the researchers have built the first detailed structural model of post-cell division NPC assembly, which will also allow them to systematically test it with targeted perturbations. Their next challenge is to create a similarly detailed model for NPC assembly during nuclear growth in the interphase phase of the cell cycle.
“Beyond the NPC, our approach of combining quantitative fluorescence live-cell imaging with existing structural data and computational modeling could also prove a useful strategy in helping to understand the assembly mechanisms of other molecular machines in the cell,” Otsuka said.
Original article by Sándor Fülöp, Communications Manager at the Max Perutz Labs Vienna
Nature 4 January 2023
10.1038/s41586-022-05528-w