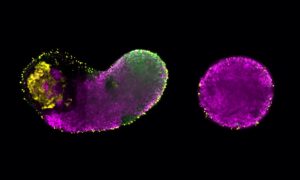
Tracing the origins of cells
EMBL scientists have developed an improved method to reconstruct a cell’s history
Researchers from the Sharpe group at EMBL Barcelona have published a method to track the developmental history of a cell using the gene editing tool CRISPR–Cas9, but without the need to create transgenic organisms.
Similar to a whole species, every cell has a lineage: its developmental history, traced back through every cell division to the first cell from which it began. Cellular lineage tracing is used to understand development. When an embryo grows, its cells divide and change their form and function, until they’ve developed into all the specialised cells an organism is made of. Tracing a cell back to its origin means understanding how it developed from a single fertilised egg cell. It also plays an important role in cancer studies, because it can be used to analyse differences between cells within a tumour that are acquired through cell division over time. Understanding these differences also leads to a better understanding of how cancer develops and progresses.
Studying genetic barcodes
One method for cellular lineage tracing uses CRISPR–Cas9: the so-called ‘genetic scissors’, which can be used to precisely cut DNA and delete a sequence or insert a new one. This makes it possible to generate mutations throughout the development of an organism and use them as lineage markers: edited pieces of DNA that lie very close to each other. This array acts as a barcode for each particular cell. By comparing barcodes, scientists can reconstruct a family tree- if the barcodes of two cells are very similar, then they are closely related. This method usually requires the creation of transgenic organisms: their genome is edited so that it contains specific DNA sequences that serve as target for the CRISPR–Cas9 machinery.
James Cotterell, Research Scientist in the Sharpe group, explains that the group identified sequences that exist naturally in the genomes of mice and zebrafish and could be used as CRISPR–Cas9 targets, so it isn’t necessary to create a transgenic organism in which you insert specific target regions into the genome. Using software written by James, the group found thousands of regions where you have arrays of possible CRISPR–Cas9 targets lying close to each other in the genome. The scientists then injected CRISPR–Cas9 machinery into the fertilised egg cell, targeting one of these arrays. A few days later, the scientists extracted DNA and found thousands of specific combinations of mutations that had accumulated over time – revealing a kind of genetic barcode that they could use to trace the cell’s lineage.
Powerful datasets
Doing cell lineage tracing without having to create transgenic organisms offers various advantages. “It simplifies the lineage tracing approach in our model systems,” says James. “We can use any zebrafish available in our facility and do lineage tracing. With the old technique, we would have had to import the specific transgenic fishline from another laboratory first.”
The lineage tracing tool is not limited to mice or zebrafish – it can be applied to any species with a published genome. “The pipeline is very pragmatic and easy,” says James. The computer program makes it possible to identify suitable target sites within an hour. This approach also enables cell lineage tracing in species where it is not possible to create transgenic specimens – in humans, for example. Scientists could use the tool to analyse human organoids – small, simplified versions of organs grown in the lab.
“Another advantage of our technique is that we can keep on adding new target sites,” says James, “so we can get more lineage information if we want to have a more detailed picture.” This can be done either by defining more target sites in a specific sequence, thus expanding the array, or by using multiple different arrays from all over the genome. The method is now available to the scientific community. It started out as a tool that was used in larger research projects in the Sharpe group, but soon became the subject of an independent publication. The group was working on a project in single-cell transcriptomics, which James describes as sequencing single cells to work out all the different cell types in an embryo. They developed the tool to combine their project with cell lineage tracing. “We were trying to include the spatial aspect as well,” says James. “By itself, a lineage tree isn’t that interesting. You need to add extra information about the cells to turn it into a powerful dataset for a given animal. Then you can determine what cell types you are looking at, where they are, and how they relate to each other in terms of development. That’s what we’re trying to do – it’s a tool for the bigger story.”


