Understanding a key player in melanoma
A new paper describes a unique mechanism of partner selectivity in transcription factors
In biology, finding the right partner is important. At the molecular level, proteins need to find ways to pair up and enable basic biological functions. Some of these proteins, specifically transcription factors, are essential for initiating, stimulating, or repressing the transcription of DNA into RNA. One feature they all have in common are characteristic DNA-binding domains that enable them to bind to specific parts of the DNA. A key family of factors are called the MIT/TFE factors. These are able to selectively partner with each other – but not with other transcription factors – to form dimers. Important cellular processes depend on this partnering being absolutely precise.
A unique principle
Researchers at EMBL have now described the mechanism that MITF, a key member of the MIT/TFE family, uses for selecting its partners. This selectivity results from the molecular structure of the protein.
Their study, published in Nucleic Acids Research, was a collaboration between the groups of Matthias Wilmanns, Head of EMBL Hamburg, and Eiríkur Steingrímsson from the University of Iceland, Reykjavík. Steingrímsson is one of the EMBL Council delegates for Iceland. He was elected Chair of the council in late November 2019 and will start as such in January 2020. Other collaborators included the group of Rainer Schindl from the Medical University of Graz, Austria.
The work of the collaboration focuses on characterising MITF and the relationship between its structure and functions. “During transcription, we deal with hundreds or even thousands of transcription factors,” Wilmanns explains. “A very important question is how transcription factors actually partner with each other. Under what conditions does transcription factor A interact with transcription factor B? This has very important implications for the processes that are going on in our body.”
Removing the stammer
Wilmanns and Steingrímsson answered these questions by observing molecules and their interactions at atomic resolution. They had already published a paper together in 2012, which characterised the naturally occurring version of MITF using structural experiments at EMBL Hamburg and functional experiments in Steingrímsson’s laboratory. During this research, they discovered a so-called stammer.
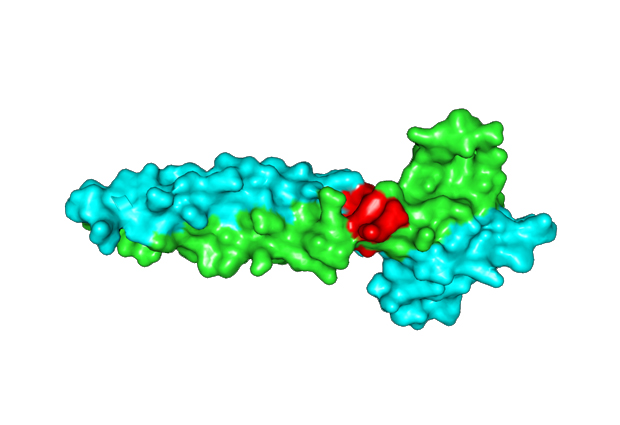
When MITF transcription factors partner with one another, the structure that emerges is called a leucine zipper, which has the form of a coiled coil. This coiled coil has periodically repeating, helical turns. If one coil gets out of reach of the other, it creates an interruption of the coiled coil in three dimensions, called a stammer.
“In this research we removed the stammer from the transcription factor,” says Wilmanns. “We showed by structural and functional data that this modified version of MITF has a different partnering pattern, interacting with other transcription factors than the original type.”
Due to its wide array of functions, it’s important to understand MITF in depth. MITF plays a key role in the development of melanoma: the most dangerous form of skin cancer. “There’s a mutation in a particular amino acid in MITF that predisposes for melanoma,” Steingrímsson explains. “People with this mutation have a five times higher chance of developing a melanoma over their lifetime. The exact function of MITF in this context is not entirely clear. It seems to be affecting the proliferation of cells.” MITF is also involved in phenotype switching: it influences the ability of cancer cells to change from proliferating to metastatic, where they travel to other sites in the body. Figuring out the exact function of MITF could be key for treating melanoma.
A history of teamwork
The collaboration between Wilmanns and Steingrímsson has a long and fruitful history. It has produced several papers, exploring over-expression or removal of natural or mutated transcription factor genes, with the goal of better understanding the relationship between structure and function, and how MITF works at the molecular level.
“I’ve been working on MITF for a long time, and I was really interested in understanding the structure of the protein,” says Steingrímsson. “I wanted to know what it looks like in 3D and how it binds to DNA.” Steingrímsson explains that an interesting fact about MITF is the high number of known mutations. By observing how these mutations affect its function, the relationship between structure and function can be understood.
This interest initiated the contact with Wilmanns. “In 2006, we had a meeting of the EMBL Council and there was this one scientist from Hamburg who was really interested in transcription factors,” says Steingrímsson. “So, I contacted him and asked if he would be interested in collaborating. That’s how we started working together.”
When asked what quality they most appreciate in their collaboration, Wilmanns and Steingrímsson both agree: “It’s straightforward and to the point.”
Wilmanns adds that the collaboration also allowed him to visit Iceland. “I think it’s really important to get to know the member states that play such an important role in the success of an organisation like EMBL,” he says.
Source articles
Pogenberg V, Ballesteros-Álvarez J, Schober R, Sigvaldadóttir I, Obarska-Kosinska A, Milewski M, Schindl R, Ögmundsdóttir MH, Steingrímsson E, Wilmanns M (2019).
Mechanism of conditional partner selectivity in MITF/TFE family transcription factors with a conserved coiled coil stammer motif.


