Unlocking the secrets of nuclear movements
EMBL researchers develop a computer model to explore the movement of nuclei in a multinuclear cell
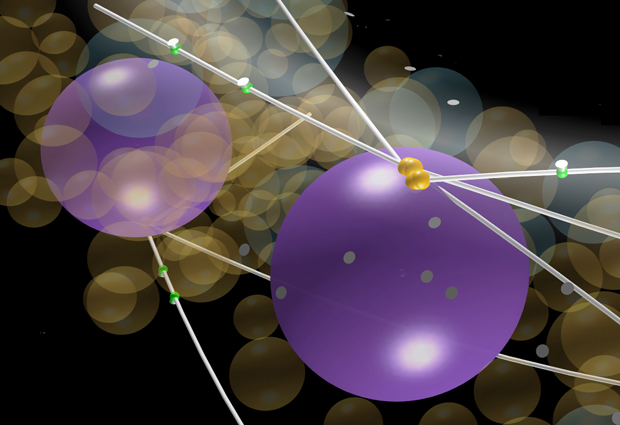
EMBL’s François Nédélec believes computer modelling can play a key role in making sense of large amounts of biological data. A new study published in Molecular Biology of the Cell shows the power of this approach.
What did you do?
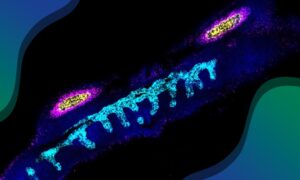
Based on the analysis of biological data obtained from different researchers, we constructed a computer model of the simple fungus Ashbya gossypii, which has cells containing many nuclei. Because the cells grow by elongating in one direction, it might be expected that the nuclei would simply move in straight lines in the same direction. However, the nuclei have protein strands called microtubules attached to them, which are pulled by dynein – a molecular motor. The nuclei can be observed making back and forth movements, and even exchanging places with one another. We wanted to know whether a system of dynein molecules pulling microtubules in random directions could explain this motion. Our model shows that it can.
Why does it matter?
This is the first time computer modelling has been used to explain the movement of multiple nuclei in a cell. We’d like to use the knowledge we’ve gained to tackle more complex problems, such as the way nuclei are positioned in human myotubes – elongated cells with multiple nuclei that are formed during muscle development. Improper positioning of the nuclei is associated with diseases such as dilated cardiomyopathy and muscular dystrophies.
Computer models have long been used to investigate problems in physics but are now increasingly important in our understanding of biological systems. They help us integrate large amounts of experimental data into a more manageable body of knowledge, allowing us to test whether our theories reflect the reality of what’s happening in cells.